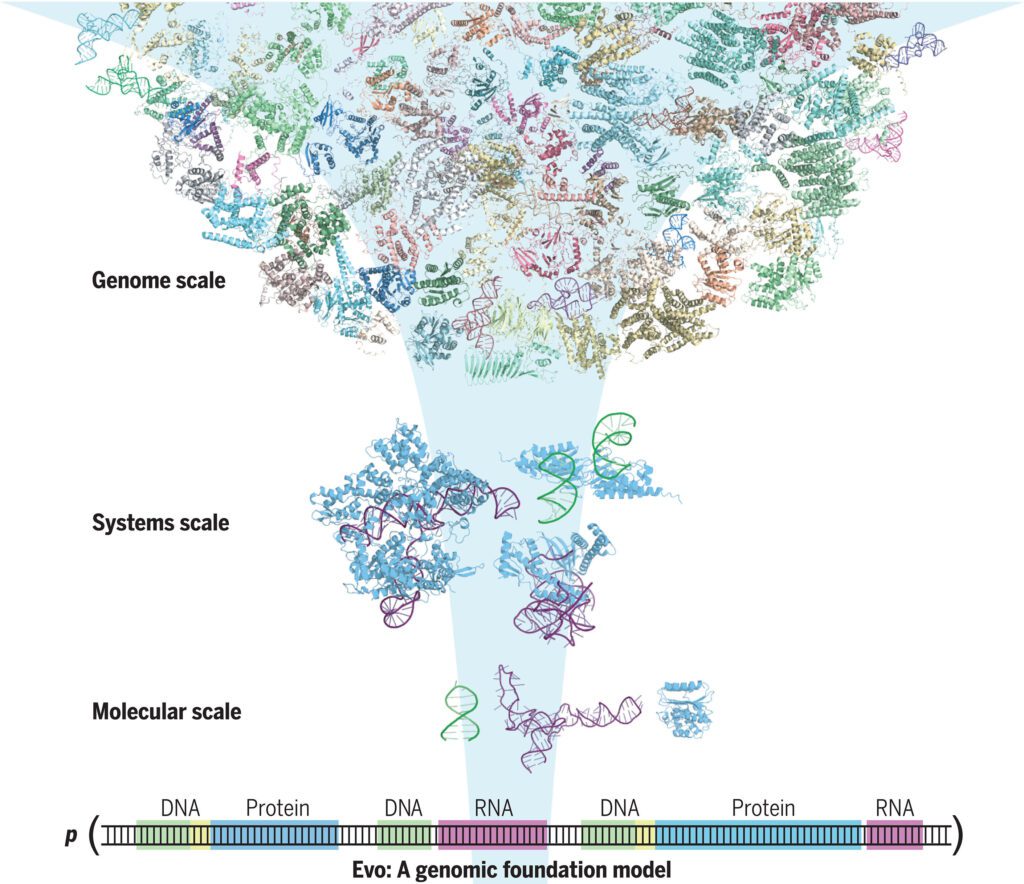
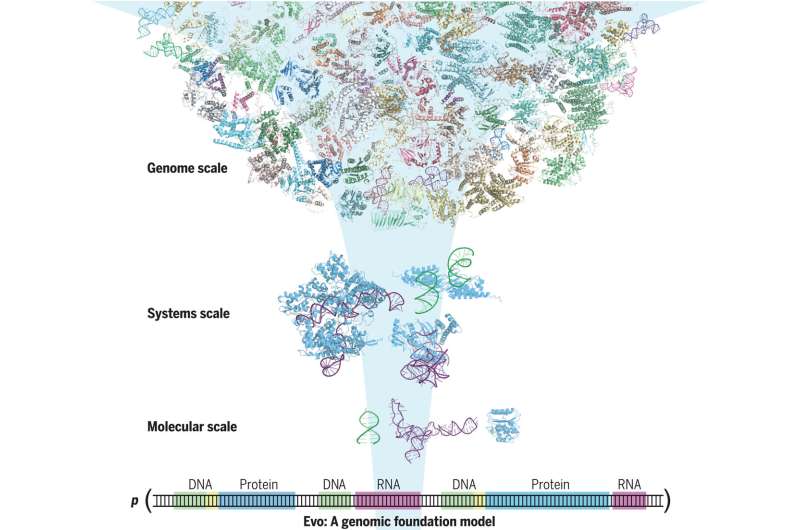
Computer scientists, bioengineers and AI specialists from the Arc Institute and Stanford University have developed an AI-based model capable of decoding and designing genetic sequences. In their paper published in the journal Science, the group describes the factors that went into designing and building their model, which they have named Evo, and list multiple possible uses for it.
Christina Theodoris, with the Gladstone Institute of Cardiovascular Disease, has published a Perspective piece in the same journal issue, outlining the work done by the researchers and suggesting the development of Evo could have major implications for medical research and treating diseases in the future.
With the advent of LLMs such as ChatGPT, computer scientists have developed tools that will likely have impacts far beyond text, image or video generation. Training a computer with vast amounts of data to help it learn opens the door to a wide range of research opportunities. For this new study, the researchers trained a computer with DNA data, which then applied it to create something new. The model, Evo, can not only decode genetic sequences but it can design new ones.
The research team trained Evo with data that showed how strands of DNA have evolved over time, and what those changes have wrought in a given organism. After receiving multiple examples of multiple types of bacteria, for example, Evo learned how their DNA operated. And that allowed the model to design a whole new microbial genome.
To give it a basic level of use, the researchers trained Evo using 2.7 million microbial genomes, which included millions of gene sequences from the types of bacteria that target viruses—a process that took four weeks. Notably, the model is able to read data down to the nucleotide level, allowing it to learn about the building blocks of the DNA strands it would soon produce.
Initial testing of the model involved asking Evo to predict what would happen with a protein given certain mutations. They compared its results with the results of experiments already conducted by humans in a lab and found them to be comparable. The difference was that Evo was able to do in minutes what took scientists years of effort.
The research team suggests that their model could soon alter the way synthetic biology is conducted.
More information:
Eric Nguyen et al, Sequence modeling and design from molecular to genome scale with Evo, Science (2024). DOI: 10.1126/science.ado9336
Christina V. Theodoris, Learning the language of DNA, Science (2024). DOI: 10.1126/science.adt3007
© 2024 Science X Network
Citation:
Evo—an AI-based model for deciphering and designing genetic sequences (2024, November 15)
retrieved 15 November 2024
from https://phys.org/news/2024-11-evo-ai-based-deciphering-genetic.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.





