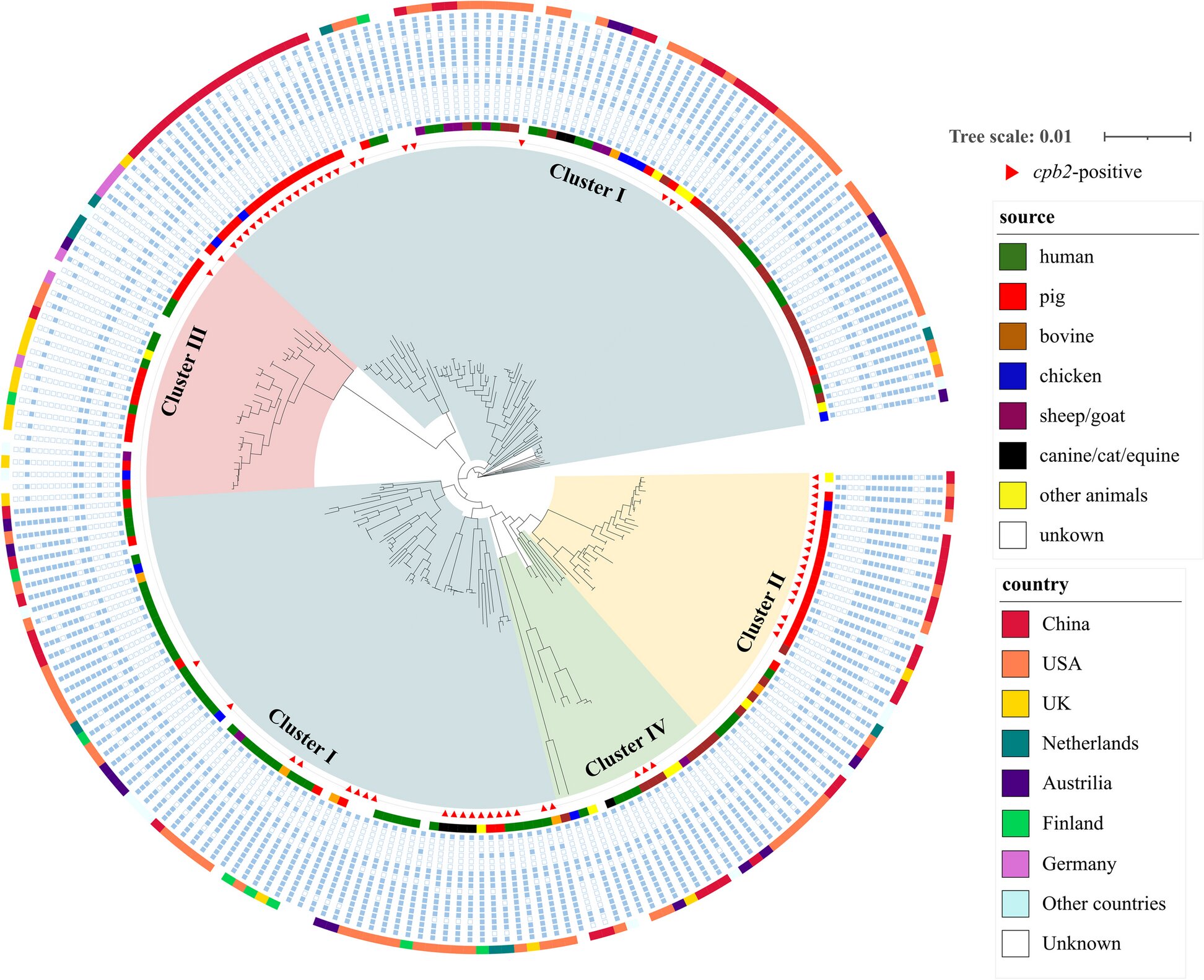
Clostridium perfringens (C. perfringens) is a widespread bacterium found in the intestines of animals and humans, known for causing severe enteric diseases. It produces over 20 toxins, with the β-2 toxin (CPB2) being linked to intestinal infections. CPB2, first identified in piglets with necrotizing enterocolitis, has since been found in multiple species. However, there is limited understanding of its genomic context and role in host adaptation. Due to these knowledge gaps, further research on the cpb2 gene and its role in pathogen evolution is essential.
The research, conducted by scientists at Northwest A&F University, was published on August 30, 2024, in the journal One Health Advances. The study aimed to investigate the genomic characteristics of C. perfringens strains carrying the cpb2 gene, with a particular focus on the potential biological function of the gene and its host specificity.
The study involved the analysis of 763 C. perfringens genomes from various hosts, revealing a high prevalence of the cpb2 gene in pig-derived strains. Interestingly, while the cpb2 gene was enriched in pigs, it did not appear to affect the evolutionary relationships of the bacterial strains. Instead, the gene is located on a conserved genetic segment within the plasmids, indicating potential horizontal transfer between strains.
The researchers also discovered that the gene is consistently located on a specific segment containing a transcriptional regulator, suggesting a mechanism for its persistence in certain hosts. Additionally, the research showed that the cpb2 gene was associated with host species-specific adaptations, particularly favoring the porcine intestinal environment. This finding suggests that C. perfringens may exploit this gene to better survive in pigs, although the gene’s function in other animals remains less clear.
Dr. Juan Wang, the lead author of the study, emphasized the importance of these findings, “Our research provides a detailed genomic analysis of the cpb2 gene, and its consistent presence in pig-derived strains suggests a key role in host adaptation. Understanding how this gene functions could lead to new insights into preventing C. perfringens infections, particularly in the livestock industry.”
The findings of this study have broad implications for public health and the livestock sector. Since C. perfringens is a major pathogen in pigs, understanding the genetic mechanisms behind its adaptation could inform new strategies for controlling infections. What’s more, these insights into the cpb2 gene could help in developing targeted interventions to prevent C. perfringens-related diseases in both animals and humans.
More information:
Ke Wu et al, Genomic insights into cpb2-positive Clostridium perfringens and the potential biological function of cpb2 gene, One Health Advances (2024). DOI: 10.1186/s44280-024-00058-8
Citation:
Beyond the toxin: Investigating the role of the cpb2 gene in the survival of Clostridium perfringens (2024, October 14)
retrieved 14 October 2024
from https://phys.org/news/2024-10-toxin-role-cpb2-gene-survival.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.