Meiotic recombination generates genetic diversity and promotes proper chromosomal segregation of parental chromosomes. This process requires a set of recombinases polymerized on single-stranded (ss) DNAs called the nucleoprotein filament to undergo homology search and strand exchange between homologous DNAs.
In Saccharomyces cerevisiae meiosis, programmed DNA double-strand breaks (DSBs) are formed by Spo11 to generate 3′-ssDNA tails. Once formed, ssDNA overhangs are rapidly bound by the abundant high-affinity ssDNA-binding protein, Replication protein A (RPA), to protect these ssDNAs from nucleolytic degradations or formation of the higher-order DNA structures.
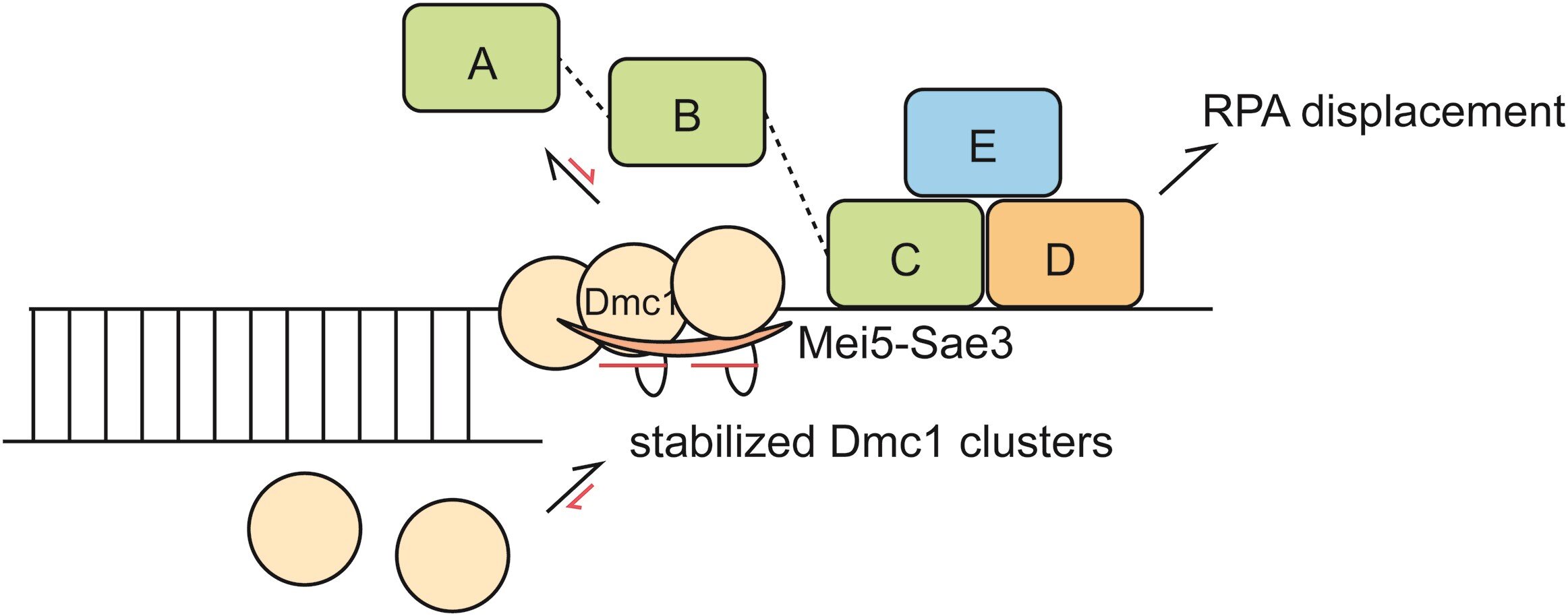
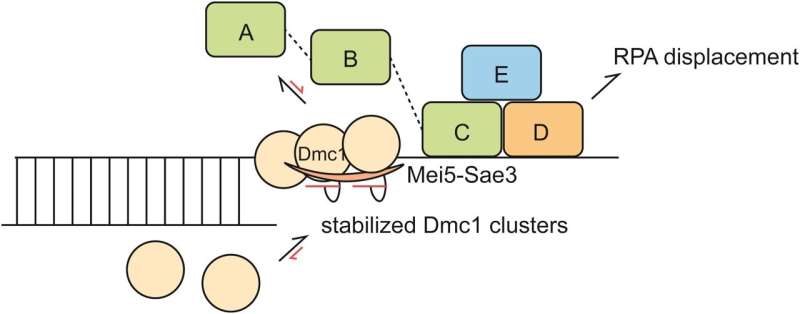
RPA-coated ssDNA substrates are distinct from bare ssDNA substrates due to RPA’s high affinity for ssDNA; therefore, the recombination mediator Mei5-Sae3 protein complex is required for the binding of recombinases onto RPA-coated ssDNA. However, the mechanistic role of Mei5-Sae3 in mediating Dmc1 activity remains unclear.
To investigate how Mei5-Sae3 stimulates Dmc1 to displace RPA and form nucleoprotein filaments, the research team, from NTU chemistry, NTU IBS, and Osaka University, utilized Biochemical protein purification techniques and single-molecule FRET and Colocalization Single-Molecule Spectroscopy (CoSMoS) techniques to capture the real-time binding of Dmc1 and dissociation of RPA on individual DNA with exceptional time resolution.
Their results are published in the journal Nucleic Acids Research.
Unlike traditional biological approaches, which mostly look at the final equilibrium products of the ensemble reactions, single-molecule methods could elucidate the contributions of individual biochemical steps from individual molecules, uncovering transient intermediate states that could give insights into how the reaction progresses.
The result showed that Mei5-Sae3 stabilized Dmc1 nucleating clusters with 2-3 molecules on naked DNA by preferentially reducing Dmc1 dissociation rates. Mei5-Sae3 also stimulated Dmc1 assembly on RPA-coated DNA.
Using GFP-labeled RPA, the co-existence of an intermediate with Dmc1 and RPA on ssDNA was observed before RPA dissociation. Moreover, the displacement efficiency of RPA depended on Dmc1 concentration, and its dependence was positively correlated to the stability of Dmc1 clusters on short ssDNA.
These findings suggest a molecular model that Mei5-Sae3 mediates Dmc1 binding on RPA-coated ssDNA by stabilizing Dmc1 nucleating clusters, thereby influencing RPA dynamics on DNA to promote RPA dissociation.
The research contributes to the first ever reported detailed molecular model for this unique mediator protein Mei5-Sae3, elucidating how a mediator-recombinase interaction can stimulate recombinase assembly on RPA-coated ssDNA.
More information:
Chin-Dian Wei et al, Mei5–Sae3 stabilizes Dmc1 nucleating clusters for efficient Dmc1 assembly on RPA-coated single-stranded DNA, Nucleic Acids Research (2024). DOI: 10.1093/nar/gkae780
Provided by
National Taiwan University
Citation:
Researchers reveal key mechanism in regulating DNA recombination (2024, October 4)
retrieved 4 October 2024
from https://phys.org/news/2024-10-reveal-key-mechanism-dna-recombination.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.