
Researchers have decoded the genomic sequence of Zygnema algae, revealing insights into the evolutionary transition from aquatic to terrestrial plant life. This breakthrough enhances our understanding of plant adaptation mechanisms and offers a basis for future studies in environmental resilience and bioenergy.
Plant life first emerged on land about 550 million years ago, and an international research team co-led by University of Nebraska–Lincoln computational biologist Yanbin Yin has cracked the genomic code of its humble beginnings, which made possible all other terrestrial life on Earth, including humans.
The team — about 50 scientists in eight countries – has generated the first genomic sequence of four strains of Zygnema algae, the closest living relatives of land plants. Their findings shed light on the ability of plants to adjust to the environment and provide a rich basis for future research.
The study was published recently in the journal Nature Genetics.
Evolutionary Insights Into Terrestrial Plant Life
“This is an evolutionary story,” said Yin, who led the research team with a scientist from Germany. “It answers the fundamental question of how the earliest land plants evolved from aquatic freshwater algae.”
Yin’s lab in the Nebraska Food for Health Center and the Department of Food Science and Technology has a long history of studying plant cell wall carbohydrates, a major component of dietary fibers for humans and farm animals; lignocelluloses for biofuel production; and natural barriers to protect crops from pathogens and environmental stresses.

All current plant life on land burst from a one-off evolutionary event known as plant terrestrialization from ancient freshwater algae. The first land plants, known as embryophyta within the clade of streptophyta, emerged on land about 550 million years ago — their arrival fundamentally changing the surface and atmosphere of the planet. They made all other terrestrial life, including humans and animals, possible by serving as an evolutionary foundation for future flora and food for fauna.
Pioneering Genomic Techniques in Algal Research
The researchers worked with four algal strains from the genus Zygnema — two from a culture collection in the United States and two from Germany. Scientists combined a range of cutting-edge DNA sequencing techniques to determine the entire genome sequences of these algae. These methods enabled scientists to generate complete genomes for these organisms at the level of whole chromosomes — something that had never been done before on this group of algae. Comparing the genomes with those of other plants and algae led to the discovery of specific overabundances of cell wall enzymes, signaling genes, and environmental response factors.
Unique Features of Zygnema Algae
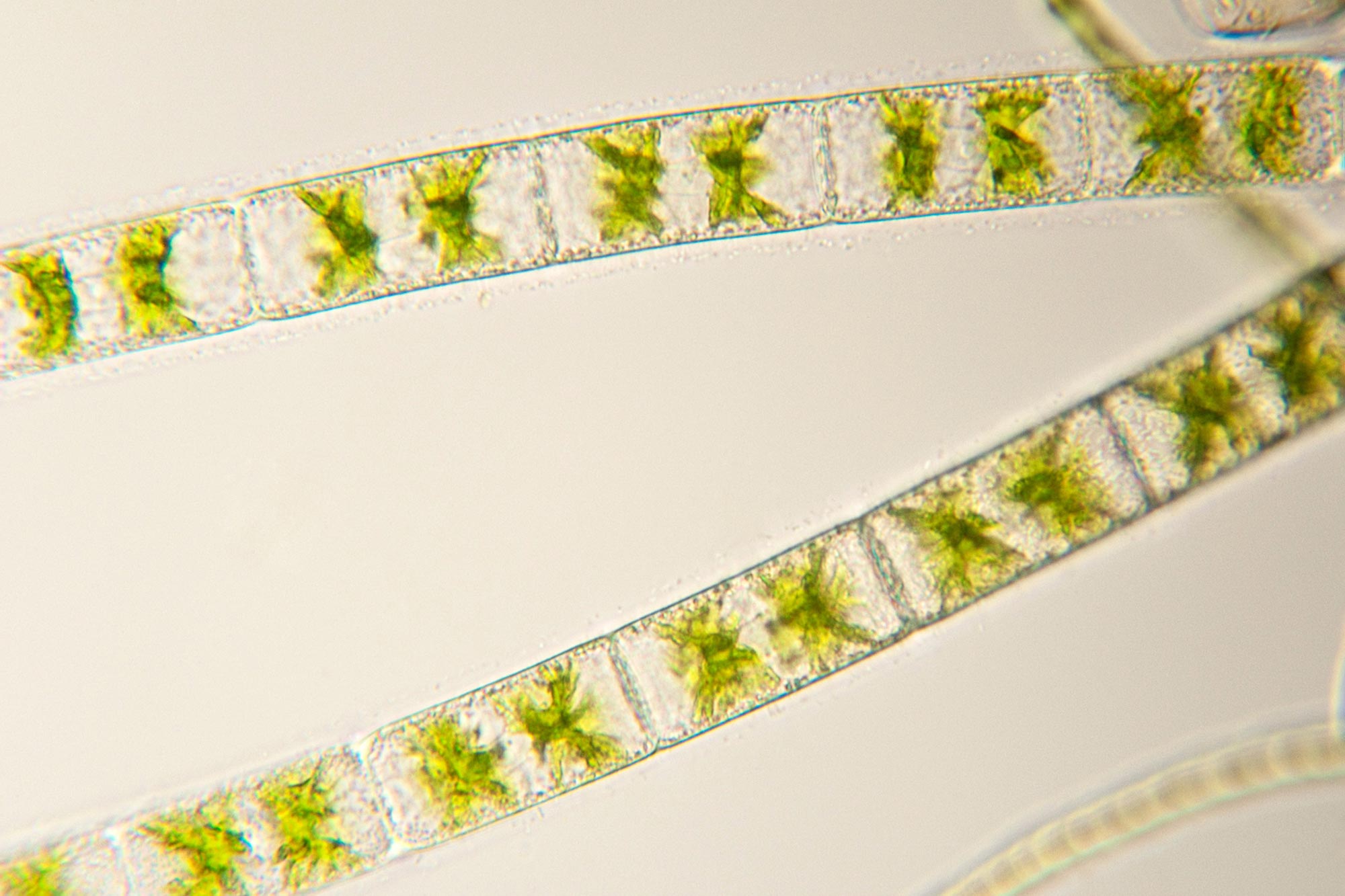
A unique feature of these algae revealed by microscopic imaging — performed at the University of Innsbruck in Austria, the Universität Hamburg in Germany and UNL’s Center for Biotechnology — is a thick and highly sticky layer of carbohydrates outside the cell walls, called the mucilage layer. Xuehuan Feng, the first author of the paper and a Husker postdoctoral research associate, developed a new and effective DNA extraction method to remove this mucilage layer for high purity and high molecular DNAs.
“It is fascinating that the genetic building blocks, whose origins predate land plants by millions of years, duplicated and diversified in the ancestors of plants and algae and, in doing so, enabled the evolution of more specialized molecular machinery,” said Iker Irisarri of the Leibniz Institute for the Analysis of Biodiversity Change and co-first author of the paper.
The team’s other co-leader, Jan de Vries of the University of Göttingen, said, “Not only do we present a valuable, high-quality resource for the entire plant scientific community, who can now explore these genome data, our analyses uncovered intricate connections between environmental responses.”
Exploring Environmental Adaptations in Algae
The four multicellular Zygnema algae belong to the class Zygnematophyceae, the closest living relatives of land plants; it is a class of freshwater and semi-terrestrial algae with more than 4,000 described species. Zygnematophyceae possess adaptations to withstand terrestrial stressors, such as desiccation, ultraviolet light, freezing, and other abiotic stresses. The key to understanding these adaptations is the genome sequences. Before this paper, genome sequences were only available for four unicellular Zygnematophyceae.
Implications for Science and Society
Yin said this research aligns with one of the National Science Foundation’s 10 Big Ideas — “Understanding the Rules of Life” — to address societal challenges, from clean water to climate resilience. The discovery also holds significance in applied sciences, such as bioenergy, water sustainability and carbon sequestration.
“Our gene network analyses reveal co-expression of genes, especially those for cell wall synthesis and remodifications that were expanded and gained in the last common ancestor of land plants and Zygnematophyceae,” Yin said. “We shed light on the deep evolutionary roots of the mechanism for balancing environmental responses and multicellular cell growth.”
The international research collaboration includes about 50 researchers from 20 research institutions in eight countries — the United States, Germany, France, Austria, Canada, China, Israel and Singapore. Other Husker researchers on the team are Chi Zhang, professor of biological sciences, and Jeffrey Mower, professor of agronomy and horticulture.
Reference: “Genomes of multicellular algal sisters to land plants illuminate signaling network evolution” by Xuehuan Feng, Jinfang Zheng, Iker Irisarri, Huihui Yu, Bo Zheng, Zahin Ali, Sophie de Vries, Jean Keller, Janine M. R. Fürst-Jansen, Armin Dadras, Jaccoline M. S. Zegers, Tim P. Rieseberg, Amra Dhabalia Ashok, Tatyana Darienko, Maaike J. Bierenbroodspot, Lydia Gramzow, Romy Petroll, Fabian B. Haas, Noe Fernandez-Pozo, Orestis Nousias, Tang Li, Elisabeth Fitzek, W. Scott Grayburn, Nina Rittmeier, Charlotte Permann, Florian Rümpler, John M. Archibald, Günter Theißen, Jeffrey P. Mower, Maike Lorenz, Henrik Buschmann, Klaus von Schwartzenberg, Lori Boston, Richard D. Hayes, Chris Daum, Kerrie Barry, Igor V. Grigoriev, Xiyin Wang, Fay-Wei Li, Stefan A. Rensing, Julius Ben Ari, Noa Keren, Assaf Mosquna, Andreas Holzinger, Pierre-Marc Delaux, Chi Zhang, Jinling Huang, Marek Mutwil, Jan de Vries and Yanbin Yin, 31 April 2024, Nature Genetics.
DOI: 10.1038/s41588-024-01737-3
Funding for UNL’s portion of the research came primarily from Yin’s NSF CAREER award, the Nebraska Tobacco Settlement Biomedical Research Enhancement Fund, the National Institutes of Health, and the U.S. departments of Agriculture and Energy.
Funding: U.S. National Science Foundation, Nebraska Tobacco Settlement Biomedical Research Development Fund, Research & Artistry Fund of Northern Illinois University, Joint Genome Institute, U.S. Department of Agriculture, NIH/National Institutes of Health, German Research Foundation, European Research Council, Austrian Science Fund, Bill and Melinda Gates Foundation, UK Foreign, Commonwealth and Development Office, Laboratoires d’Excellence