A unique method of monitoring river health has uncovered an army of tiny organisms fighting to protect the Brisbane River.
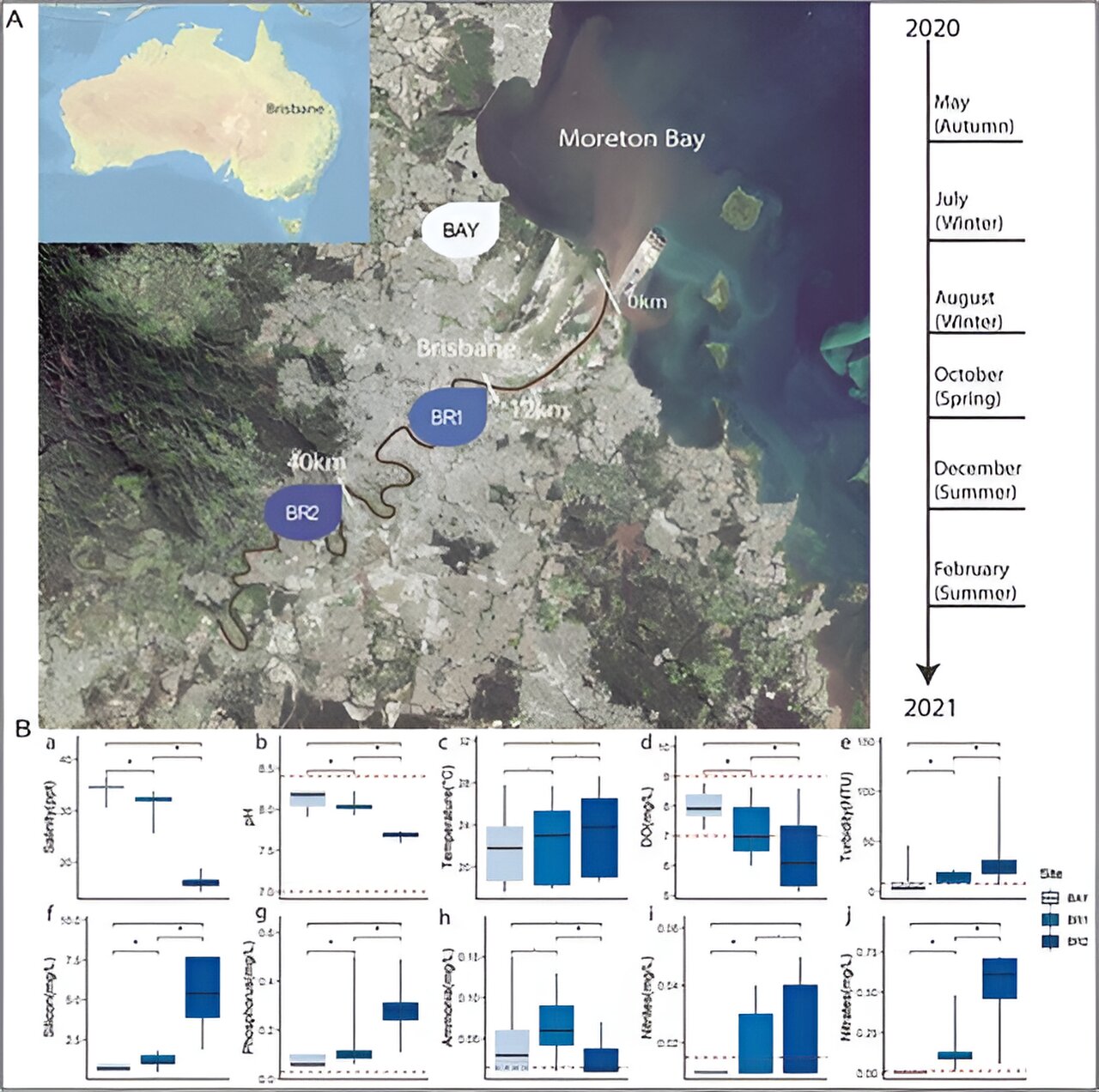
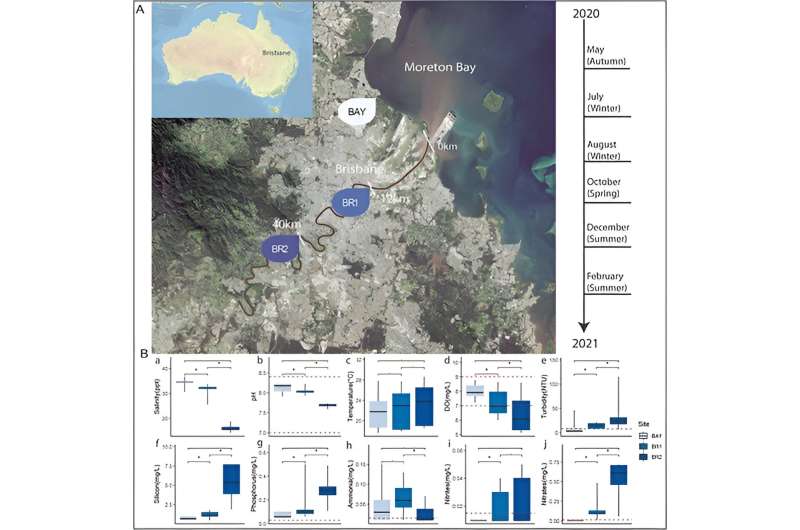
University of Queensland Ph.D. candidate Apoorva Prabhu and Honorary Associate Professor Chris Rinke led a team that sampled, sequenced and evaluated the DNA of thousands of microorganisms in 3 parts of the Brisbane River over a 12-month period.
The team’s research is published in the journal ISME Communications.
Almost 2,500 different types of microbial genomes were sampled and identified—many belonging to previously unexplored species—forming the largest ever microbial genome database for a subtropical estuary.
“The full extent of microbial diversity in the Brisbane River had been unknown, until now,” Mrs. Prabhu said. “Microbes are a handy indicator of the health of our waterways. They’re efficient nutrient recyclers, processing carbon, nitrogen, phosphorous and sulfur.
“Unfortunately, at our Brisbane River collection sites at Indooroopilly, Eagle Farm and Shorncliffe in Moreton Bay we found a high number of microbes which feed on nutrients like nitrates or phosphorous, indicating significant pollution from human activity such as agricultural runoff.
“Specifically, a novel bacterium which we named Hypereutrophica brisbanensis proved to be an important indicator for elevated levels of nitrates and phosphorous. Whenever we encountered this bacterium, nutrient levels in the river were through the roof, and sadly the reality is while these microbes remain well-fed, the river is suffering.”
The team used advanced metagenomics—a method designed to study entire communities of microorganisms—alongside machine learning techniques to identify a substantial number of new microbes.
“Remarkably, more than 80 percent of the microorganisms we found were entirely new to science, highlighting a vast and previously unexplored diversity within the lower Brisbane River,” Mrs. Prabhu said. “This knowledge is vital for developing effective environmental monitoring and management strategies, as it specifically tells us what the most pressing issues are, pollution-wise, facing our waterways.”
Dr. Rinke said the study sets a benchmark for microbial ecology research in estuarine environments, particularly in subtropical and tropical regions such as Brisbane.
“The integration of metagenomics and machine learning presents a robust framework for exploring microbial communities and their ecological functions,” Dr. Rinke said.
“Future research can build on these findings to further explain the complex interactions within microbial ecosystems and their responses to environmental changes. Our next aim is to focus on the impact of floods on microbial communities. I am sure the Brisbane River has many more secrets lurking in its muddy waters.”
The team would like to acknowledge the contribution of researchers from UQ’s School of Chemistry and Molecular Biosciences and QUT’s Centre for Microbiome Research.
More information:
Apoorva Prabhu et al, Machine learning and metagenomics identifies uncharacterized taxa inferred to drive biogeochemical cycles in a subtropical hypereutrophic estuary, ISME Communications (2024). DOI: 10.1093/ismeco/ycae067
Provided by
University of Queensland
Citation:
Study reveals the microbes vital to a healthy Brisbane River (2024, June 27)
retrieved 27 June 2024
from https://phys.org/news/2024-06-reveals-microbes-vital-healthy-brisbane.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.