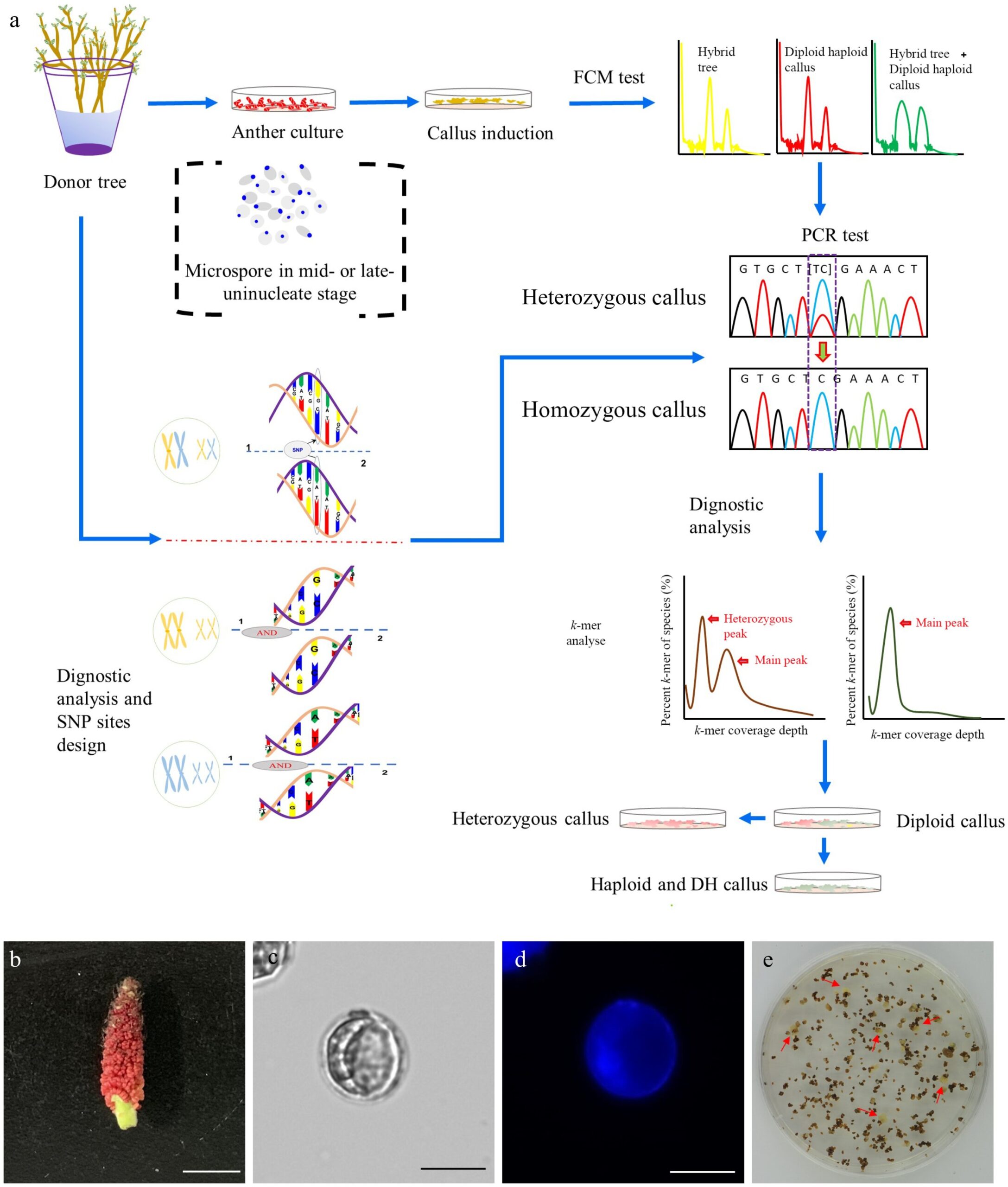
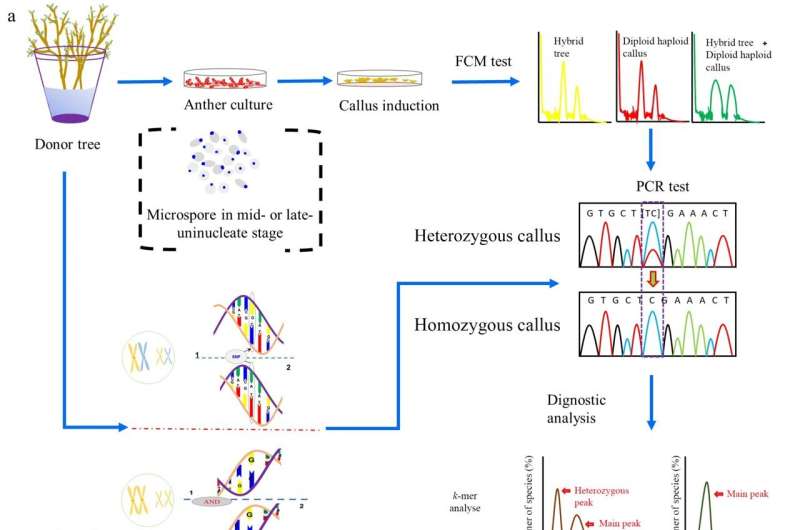
A research team has successfully assembled a nearly gap-free, telomere-to-telomere (T2T) genome of Populus ussuriensis, filling gaps present in the P. trichocarpa genome. Leveraging long-read sequencing, the team identified and annotated centromere regions in all double haploid (DH) genome chromosomes, providing a first for poplars. With 34,953 protein-coding genes, this genome surpasses P. trichocarpa by 465 genes.
The T2T P. ussuriensis genome enhances understanding of poplar genome structure and functions, aiding in poplar evolutionary studies. The assembly’s high collinearity with P. trichocarpa facilitates comparative genomics, epigenetic research, and reproductive biology studies, marking a significant contribution to the field.
Poplars, renowned for their relatively short life cycle and wide adaptability, have become pivotal in various industries and reforestation efforts due to their versatile applications and pioneer tree characteristics. Despite their significance, achieving high-quality genome assemblies in poplars, especially Populus trichocarpa, remains challenging due to their high heterozygosity and highly repetitive sequences in the genomes.
Recent advancements in sequencing technologies have improved assembly quality, yet high genomic heterozygosity persists as a barrier. The development of homozygous lines offers a potential solution, but the long juvenile periods of woody plants pose practical challenges. While DH lines have been utilized in crop genome sequencing, their application in forest trees remains limited.
Addressing this gap and developing efficient methods for inducing haploid plants or DH lines in poplars could revolutionize genomic research in these dioecious trees, enabling more accurate and comprehensive genome assemblies for enhanced understanding of their biology and environmental adaptability.
A study published in Forestry Research reveals a high-quality poplar genome with annotated centromeres and telomeres, facilitating molecular analyses and comparative genomics.
The study initiated haploid callus induction from P. ussuriensis anthers, identified haploid and DH calli via high-throughput screening, and conducted whole-genome resequencing for SNP identification. DH15 line was selected, and k-mer analysis estimated its homozygous origin and genomic size. A gap-free reference genome was constructed for DH15 using PacBio HiFi reads and Hi-C data, featuring 19 fully assembled chromosomes, annotated telomeres, and centromeres.
The genome exhibited high completeness and quality, surpassing previous P. trichocarpa assemblies. Telomeres displayed distinct lengths across chromosomes, while 19 centromeres with varying lengths and structures were identified. Comparative analysis revealed close phylogenetic relationships among Populus species. The DH15 genome, with 34,953 protein-coding genes, demonstrated a diverse array of repetitive elements. Notably, gene family analysis identified specific gene families enriched in phosphate metabolic processes. Comparison with other Populus genomes highlighted DH15’s superior contiguity and completeness, particularly in centromere regions, unveiling new genes and transcripts.
According to the study’s senior researcher, Su Chen, “This refined genome assembly will be highly instrumental in molecular analyses of gene functions in poplar trees and enable comparative genomic studies across different poplar species. It serves as a solid foundation for future research on the poplar and other plant genomes.”
In summary, this study achieved a near-gapless assembly of a highly contiguous poplar genome, DH15, enabling comprehensive analyses of centromeric regions and gene functions, which will drive advances in poplar genomics, comparative studies, and molecular breeding strategies. Looking ahead, this refined genome assembly will serve as a crucial resource for understanding poplar biology and accelerating practical applications in forestry and biotechnology.
More information:
Wenxuan Liu et al, A nearly gapless, highly contiguous reference genome for a doubled haploid line of Populus ussuriensis, enabling advanced genomic studies, Forestry Research (2024). DOI: 10.48130/forres-0024-0016
Provided by
Maximum Academic Press
Citation:
Advancing poplar genomics: Nearly gap-free genome assembly unveils new insights and applications (2024, May 29)
retrieved 29 May 2024
from https://phys.org/news/2024-05-advancing-poplar-genomics-gap-free.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.

