A study has tapped into the peach genome, creating large-scale gene co-expression networks (GCNs) that predict gene functions and streamline the peach breeding process. This innovative approach addresses the complex task of identifying genes linked to desirable breeding traits in peaches.
Peach genetics faces challenges such as long intergeneration times and limited genetic transformation systems, making gene identification and validation complex. Traditional approaches often fall short in predicting gene functions due to these limitations.
Gene co-expression networks (GCNs), which map the relationships between gene expressions, offer a promising solution. By leveraging the “guilty-by-association” principle, GCNs can predict gene functions based on shared expression patterns. Due to these challenges, there is a pressing need for innovative tools like GCNs to enhance our understanding of peach genetics.
Researchers from the Institut de Recerca i Tecnologia Agroalimentàries (IRTA) and Centre de Recerca en Agrigenòmica (CRAG) published a study in the journal Horticulture Research on January 2, 2024. The study introduces a new tool that leverages GCNs to predict gene functions in peaches, potentially revolutionizing fruit breeding.
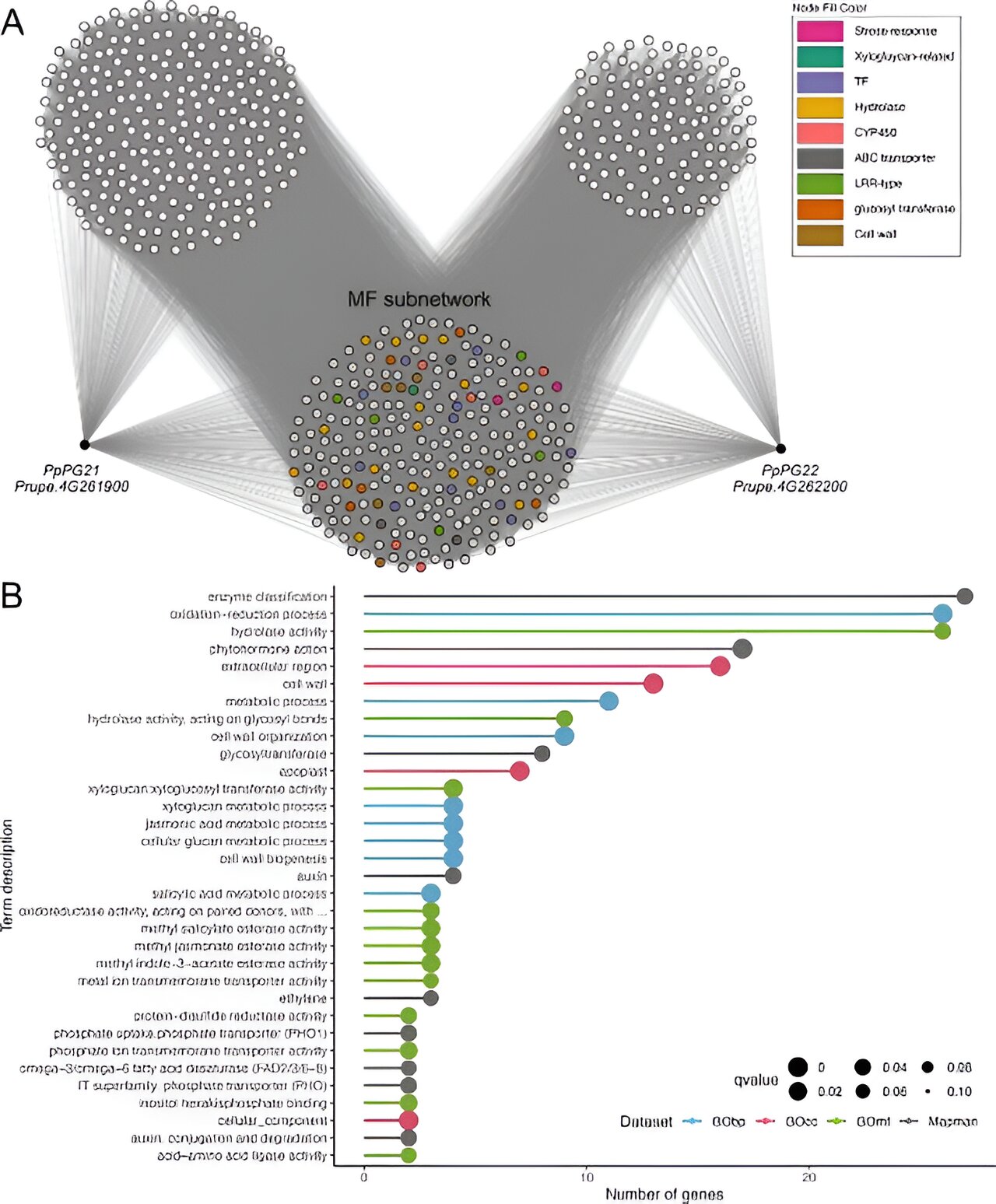
The study created four GCNs from 604 Illumina RNA-Seq libraries and evaluated their performance using a machine-learning algorithm based on the “guilty-by-association” principle. Among the networks, COO300 showed the best performance, encompassing 21,956 genes. To validate COO300, researchers conducted two case studies.
In the first case study, genes PpPG21 and PpPG22, involved in peach fruit softening, were analyzed. The co-expressed genes formed the melting flesh (MF) network, which was enriched with terms related to cell wall organization and phytohormone signaling. The MF network included genes associated with cell wall disassembly and ripening-related phytohormones, demonstrating COO300’s ability to predict gene functions accurately.
In the second case study, the transcription factor PpMYB10.1, which regulates anthocyanin accumulation, was examined. The fruit color (FC) network identified genes involved in anthocyanin metabolism and regulation. By comparing the FC network with grapevine orthologs, researchers identified conserved regulatory networks, further validating COO300’s predictive capabilities.
Dr. Iban Eduardo, a leading researcher at CRAG, stated, “The development of the COO300 network represents a significant breakthrough in peach genetics. By accurately predicting gene functions, this tool not only enhances our understanding of peach biology but also paves the way for more targeted and efficient breeding strategies.”
The study offers breeders a powerful tool to enhance peach varieties. By decoding gene functions, the PeachGCN v1.0 and its scripts, accessible to all, is expected to drive advancements in flavor, longevity, and nutrition of peaches. This genomic insight is a harbinger of change, signaling a shift towards data-driven breeding in agriculture.
More information:
Felipe Pérez de los Cobos et al, Exploring large-scale gene coexpression networks in peach (Prunus persica L.): a new tool for predicting gene function, Horticulture Research (2024). DOI: 10.1093/hr/uhad294
Provided by
NanJing Agricultural University
Citation:
Peach perfection: Advanced gene networks reveal fruit traits (2024, May 20)
retrieved 20 May 2024
from https://phys.org/news/2024-05-peach-advanced-gene-networks-reveal.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.