Circular ribonucleic acids (circRNAs) are a promising platform for gene expression studies as a stable and prevalent ribonucleic acid in eukaryotic cells, which arise from back-splicing. In a new report now published in Nature Biotechnology, Robert Chen and a team of interdisciplinary researchers at Stanford University, California, U.S., developed a systematic approach to rapidly assemble and test features affecting protein production based on synthetic circular RNAs. The team maximized translation of the circRNA by optimizing fine elements to implement design principles to improve circular RNA yield by several hundred-fold. The outcomes facilitated an increased translation of the RNA of interest, when compared to messenger RNA (mRNA) levels, to provide durable translation in vivo.
Developing circular RNA (circRNA) in the lab
Therapeutics based on ribonucleic acids span across messenger RNA (mRNA), small interfering RNAs (siRNA) and microRNAs (miRNA) with expansion into modern medicine including small molecules, biologics and cell therapeutics. For example, the lately popular mRNA vaccines can be designed in the lab and developed at a rapid pace to respond to evolving and urgent medical crises. Coding RNAs can be circularized into circRNAs to extend the duration of protein translation, based on RNA molecules that covalently join head-to-tail. Bioengineers have also advanced the synthesis of circular long transcripts into circRNAs. However, the fundamental mechanisms of initiating translation to form circular RNA or messenger RNA differ due to the lack of a 7-methylguanylate (M7G) cap on the circular RNAs. As a result of this, researchers need to thoroughly examine the principles of circular RNA translation to build better therapies and potentially surpass the translational capacities of mRNA. To examine this aspect, the team developed a modular high-throughput platform to build and test synthetic circular RNAs for optimized translation and improved protein yields.
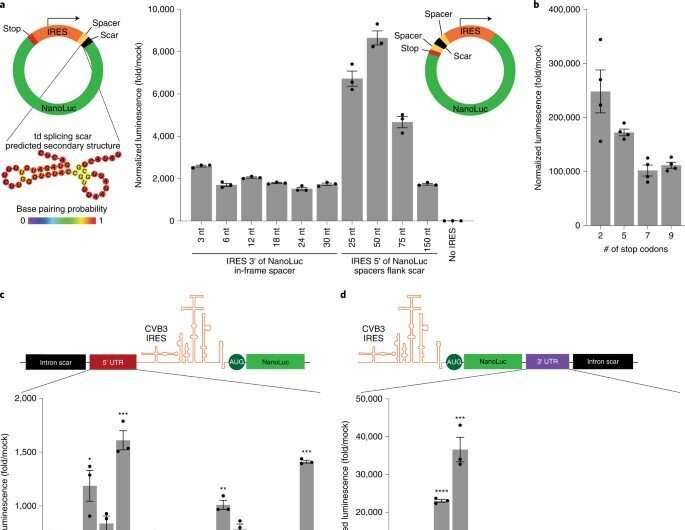
A modular circRNA assembly platform
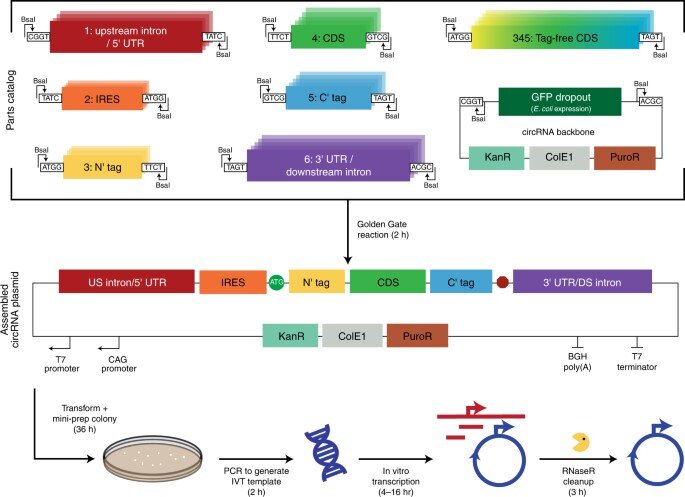
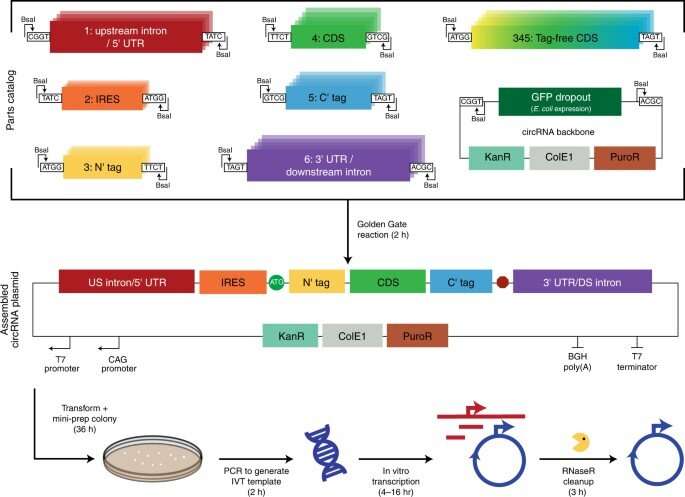
The scientists developed a modular cloning platform made of a set of parts compatible with Golden Gate and Gibson cloning to allow higher-throughput testing of circRNAs. Using the platform, they determined how specific aspects of circular RNA design affected its translation. For instance, the team had previously shown how circular RNA triggered immune responses can be avoided in vivo by modifying the molecules with m6A. However, researchers must still understand the impact of this step on circular RNA translation. To address this, Chen and the team used their cloning platform and incorporated m6A. When compared to unmodified circRNAs, those containing 5% m6A showed equal translation after transfection or electroporation in vitro. The scientists thus experimentally gauged the impact of the modification on circular RNA stability.

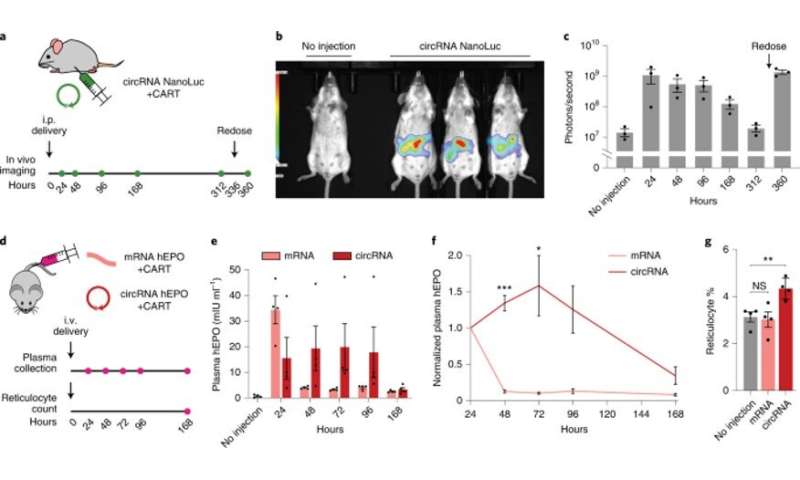
Engineered circRNAs demonstrate more durable translation and functional activity in vivo. (a) CircRNA with 5% m6A incorporation encoding NanoLuc was synthesized with the following optimizations: upstream IRES topology, 5′ PABP spacer, HBA1 3′ UTR and HRV-B3 IRES with proximal loop Apt-eIF4G insertion. CircRNAs were formulated for intraperitoneal delivery in mice using CARTs. Expression was assayed using an optical imaging system after intraperitoneal injections of the fluorofurimazine substrate at the indicated timepoints. At 336 hours (14 days) after circRNA NanoLuc administration, mice were redosed. (b) In vivo luminescence image of an untreated mouse (left) versus mice receiving circRNA NanoLuc (right) at 24 hours after dosing. (c) Quantification of luminescence per mouse at different timepoints after circRNA NanoLuc administration. Redosing was performed at 336 hours (14 days). Data are mean ± s.e.m. for n = 3 animals per condition. (d) CircRNA with 5% m6A incorporation encoding hEPO was synthesized with the following optimizations: upstream IRES topology, 5′ PABP spacer, HBA1 3′ UTR and HRV-B3 IRES with proximal loop Apt-eIF4G insertion. mRNA-encoding hEPO was synthesized with CleanCap reagent, 100% N1Ψ incorporation and a 120-nt poly(A) tail. Equimolar doses of circRNA and mRNA were formulated for intravenous delivery in mice using CARTs. Plasma hEPO was measured by ELISA in one cohort at the indicated timepoints. Reticulocytes were counted in a separate cohort at 168 hours (7 days). (e) Quantification of plasma hEPO at different timepoints after circRNA hEPO or mRNA hEPO administration. Data are mean ± s.e.m. for n = 4 animals per condition. (f) Plasma hEPO expression normalized to the 24-hour level of each mouse at different timepoints after circRNA hEPO or mRNA hEPO administration. Data are mean ± s.e.m. for n = 4 animals per condition. *P = 0.0487 and ***P = 0.0001 by unpaired two-sided t-test with Bonferroni correction compared to mRNA. g, Reticulocyte percentage among red blood cells at 168 hours after circRNA hEPO or mRNA hEPO administration. Data are mean ± s.e.m. for n = 4 animals per condition. **P = 0.0080 by unpaired two-sided t-test. NS, not significant. Credit: Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01393-0 
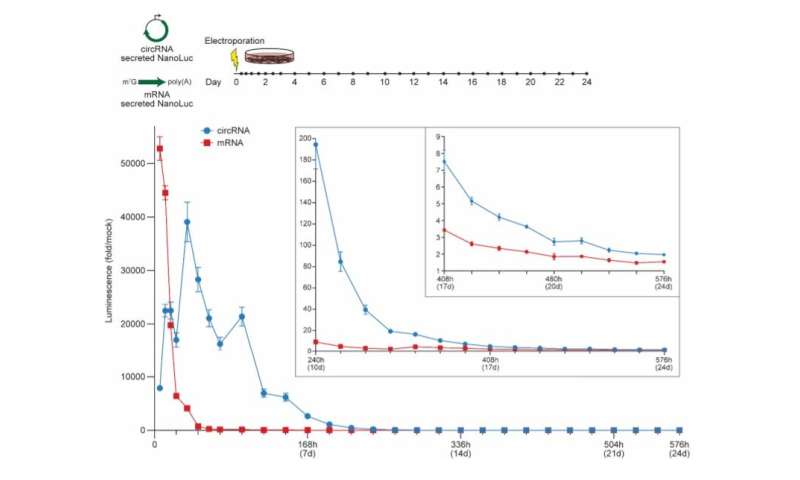
CircRNA exhibits more durable translation than mRNA in vitro. NanoLuc activity in supernatant after electroporation of HeLa cells with circRNA or mRNA encoding secreted NanoLuc. CircRNA was synthesized with 5% m6A incorporation and the HRV-B3 IRES. mRNA was synthesized with CleanCap reagent, 100% N1Ψ incorporation, and a 120 nt poly(A) tail. At the indicated hours (h) and days (d) post-electroporation, media was harvested to assay secreted NanoLuc and replaced. NanoLuc activity was divided by values from mock electroporation. Data are mean ± SEM for n=3 biological replicates. Credit: Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01393-0
Uncovering the dynamics of circRNA for strong translation outcomes
To uncover the principles underlying circRNA vector topology necessary for strong translation, the researchers began synthesizing circRNAs to generate variants with peptides encoded by the process. Based on the outcomes, the team showed that increasing the spacer-length was non-beneficial for translation. Next, they showed how the 5′ and 3′ untranslated regions could improve circRNA translation. The researchers also conducted a series of experiments to examine circRNA optimization and then compared them in a single experiment. They showed how the changes progressively increased the expression of circRNA without compromising the RNA yield or efficiency of circularization. They also showed how the kinetics of circRNA and mRNA translation significantly differed, where circRNA took more than 24-hours to reach its maximum translation length, far exceeding the translation duration of mRNA. They then combined the series of circRNA optimizations to test their expression in vivo. To deliver the RNAs, the team formulated them with charge-altering releasable transporters (CARTs) or cationic molecules mediating mRNA expression in mouse models. The outcomes showed how the engineered circRNAs could be expressed at strengths similar to modified RNAs in vivo, albeit with greater duration.
Outlook
In this way, Robert Chen and colleagues showed how RNA circularization has great potential to transform RNA-based medicines by extending the durability of relatively highly transient molecules. Given the fundamental differences between the mechanisms of circRNA and mRNA translation, the existing knowledge of maximizing mRNA translation did not necessarily translate to circRNAs. To facilitate this study, the team created a circRNA modular cloning platform to test numerous sequence variations and optimizations of multiple parameters. Using the platform, they identified several approaches to improve protein translation from circRNAs, with applications to broadly engineer RNAs, to produce more protein than mRNAs in vitro, and exhibit greater durability of its translation in vivo and in vitro. The scientists systematically dissected the elements regulating circRNA translation to then optimize the regions of interest for increased circRNA protein yields for durable protein production in vivo.
Researchers develop interactive database for translatable circular RNAs based on multi-omics evidence
Robert Chen et al, Engineering circular RNA for enhanced protein production, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01393-0
Chang-you Chen et al, Initiation of Protein Synthesis by the Eukaryotic Translational Apparatus on Circular RNAs, Science (2006). DOI: 10.1126/science.7536344
© 2022 Science X Network
Citation:
Engineering circular ribonucleic acids (circRNAs) for improved protein production (2022, August 15)
retrieved 15 August 2022
from https://phys.org/news/2022-08-circular-ribonucleic-acids-circrnas-protein.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.