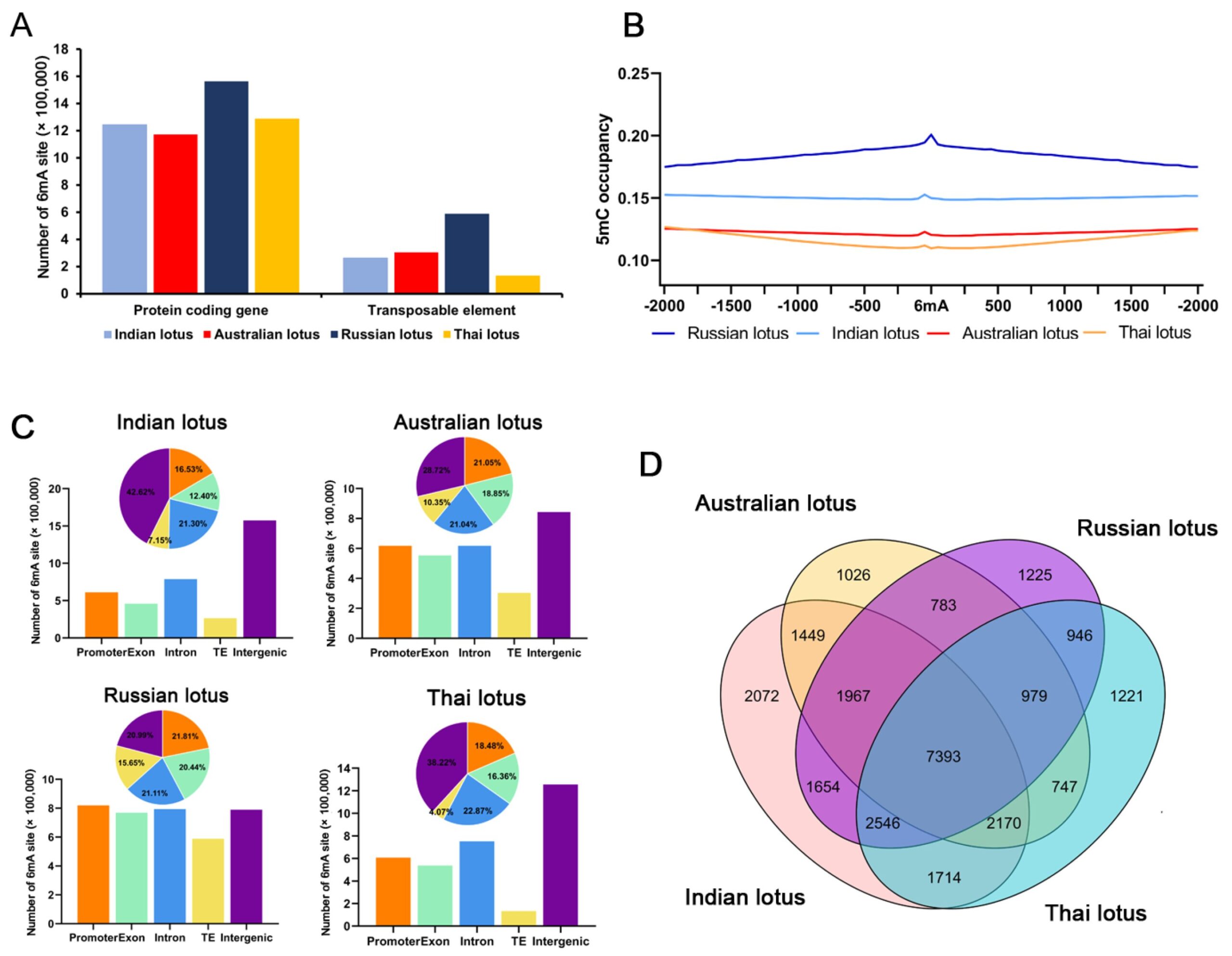
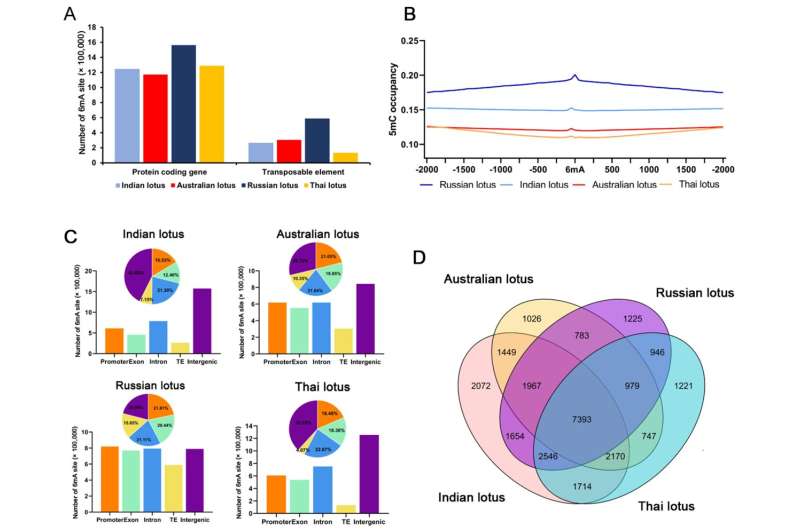
N6-methyladenine (6mA) is as an important epigenetic modification in eukaryotes. Although 6mA was discovered at the same time as 5-methylcytosine (5mC), it has only recently received attention in eukaryotes, mainly due to the limitations of detection technology.
Methylation of genomic DNA plays a critical role in gene regulation and genome stability in eukaryotes. However, it is unclear how 6mA methylation on genes evolves across the genome during species divergence and divergence of different duplicate genes.
To understand the evolution of 6mA methylation, researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences (CAS) investigated the differences in 6mA methylation within four wild species of Nelumbo nucifera and the patterns of 6mA methylation between N. nucifera, Arabidopsis thaliana, and Oryza sativa orthologs. The study, entitled “6mA DNA methylation on genes in plants is associated with gene complexity, expression and duplication,” was published in the journal Plants.
Four high-quality lotus reference genomes were sequenced and assembled into pseudochromosome using Oxford Nanopore Technologies long-read sequencing.
Distribution analysis detects no similarity between 6mA sites and the widely studied 5mC methylation sites in lotus. Consistently, 6mA sites are enriched at the start sites, positively correlated with gene expression, and preferentially retained in highly, and widely, expressed genes with long lengths among distantly related plants.
Among different duplicated genes, 6mA modifications are more likely to be retained in whole-genome duplications than in locally duplicated genes, during the long-term evolution of plant species.
More information:
Yue Zhang et al, 6mA DNA Methylation on Genes in Plants Is Associated with Gene Complexity, Expression and Duplication, Plants (2023). DOI: 10.3390/plants12101949
Provided by
Chinese Academy of Sciences
Citation:
Scientists reveal genomic distribution and evolutionary patterns of 6mA modifications in plants (2023, July 4)
retrieved 4 July 2023
from https://phys.org/news/2023-07-scientists-reveal-genomic-evolutionary-patterns.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.