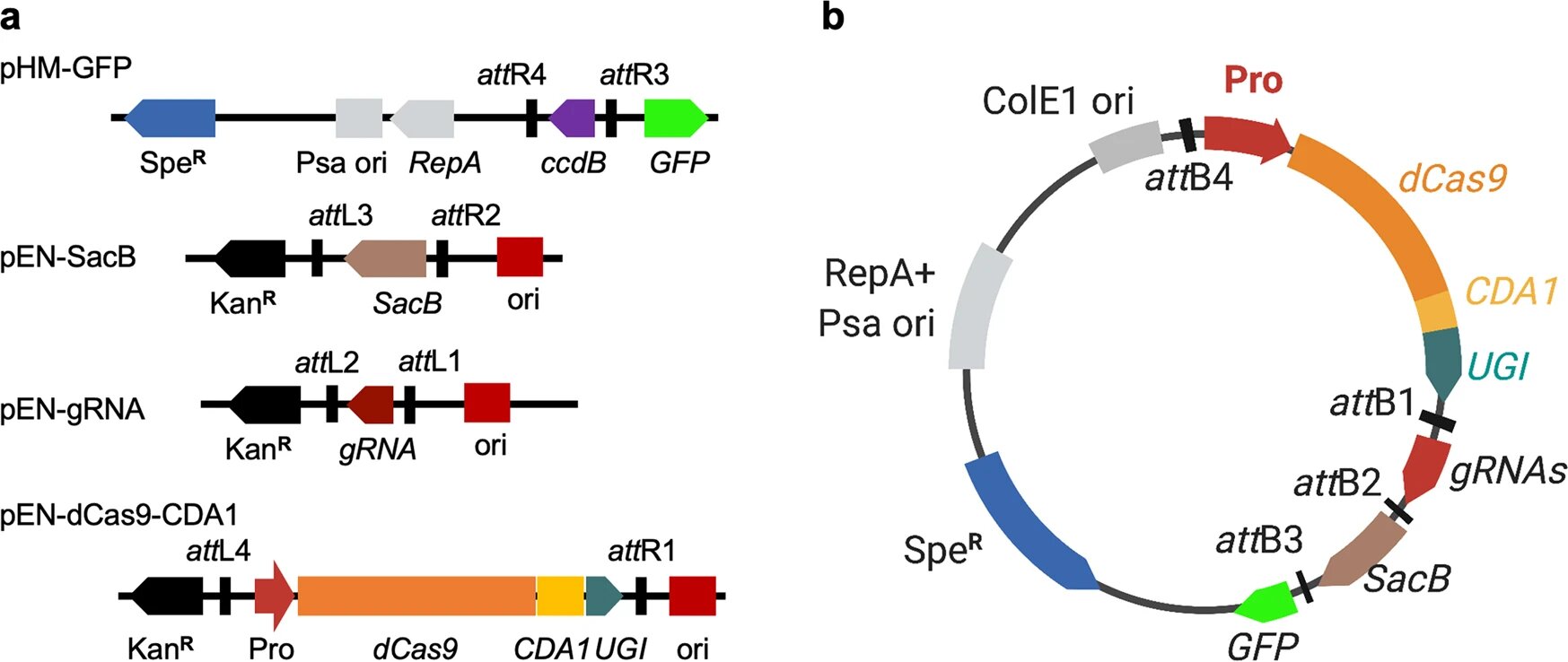
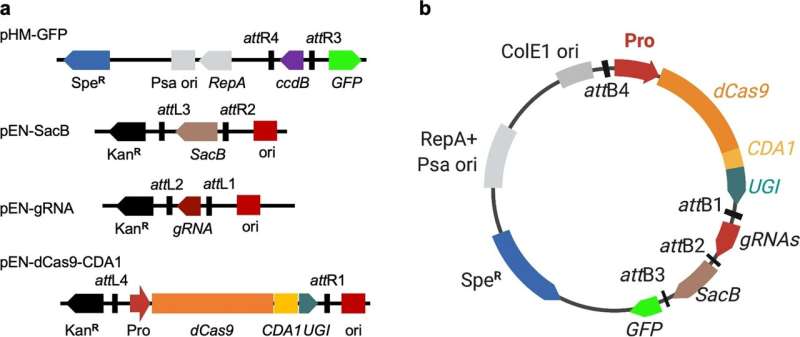
As global food insecurity climbed to a perilous high in 2022, scientists ramped up their efforts to perfect best practices for protecting the yields of major crops that are essential in combating this issue. And, while rice makes up a small portion of Missouri’s annual harvest, it—along with corn and soybeans—are key staples that help address food insecurity not only in the United States, but across the world.
In a recent study that examined how diseases function in rice crops, University of Missouri researchers might have found critical answers.
In this study, Bing Yang, a plant biology professor in the MU College of Agriculture, Food and Natural Resources and the Donald Danforth Plant Science Center used genome editing as a tool to identify problematic pathogens present in certain bacteria that lead to prolific infections in rice crops. His research helps scientists understand how these pathogens function and, thus, can determine how to guard against widespread infections that destroy yields.
This research provides insight into the host-pathogen relationship, allowing scientists to better genetically engineer plants to survive crop diseases.
“Based on the advances in scientific understanding made during this study, we are now able to develop strategies to engineer host-resistance against the bacteria,” Yang said. “That’s how we can support plant resistance in general.”
First, the research team discovered a way to “knock out” genes. When specific genes are knocked out of the bacteria, it allows scientists to better understand the functions of those specific genes. Researchers then tested for infectious properties—something that has historically been a labor-intensive process.
“This research allows us to better understand which bacteria hold pathogenic qualities and how those qualities correspond to infections in certain species of plants,” Yang said. “Ultimately, these advances in gene editing help us modify the genome of crops, in this case rice, in ways that build resistance that protect them from diseases.”
Using a revolutionary gene-editing technique called CRISPR—a method where scientists edit genes by cutting DNA and then letting it repair naturally—Yang and his team edited a sample of bacteria with the goal of determining exactly which genes had pathogenic qualities that would infect proteins in the genome of the rice crop.
Notably, Yang’s method revolutionizes a process known as homologous recombination, which has been known to be ineffective and time-consuming.
“My long-term research goal is a better understanding of the biology of diseases and plant biology and the process of using advanced technology to engineer disease resistance,” Yang said. “I also want to engineer the product so it’s more nutritious and of a higher quantity while increasing a season’s yield and reducing the yield loss.”
As a scholar of plant pathology for more than 15 years, Yang said that bacteria, including those with symbiotic (beneficial) and pathogenic qualities, are essential in maintaining life and the health of our ecosystems.
“Efficient CRISPR-Cas9 based cytosine base editors for phytopathogenic bacteria,” was published in Communications Biology.
More information:
Chenhao Li et al, Efficient CRISPR-Cas9 based cytosine base editors for phytopathogenic bacteria, Communications Biology (2023). DOI: 10.1038/s42003-023-04451-8
Provided by
University of Missouri
Citation:
Using a gene-editing tool to improve productivity in rice crops (2023, June 1)
retrieved 1 June 2023
from https://phys.org/news/2023-06-gene-editing-tool-productivity-rice-crops.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.