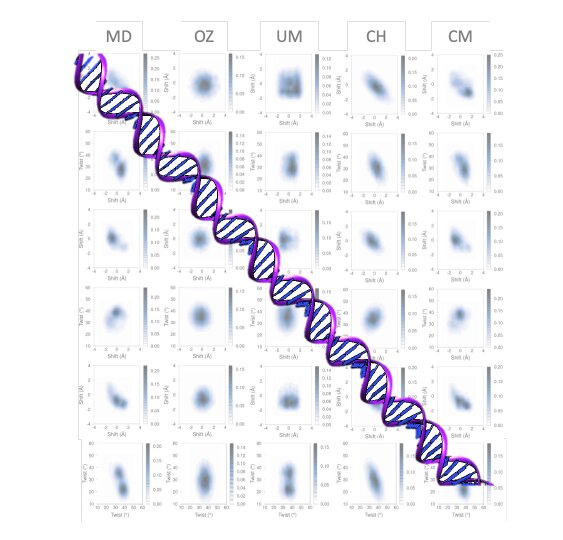
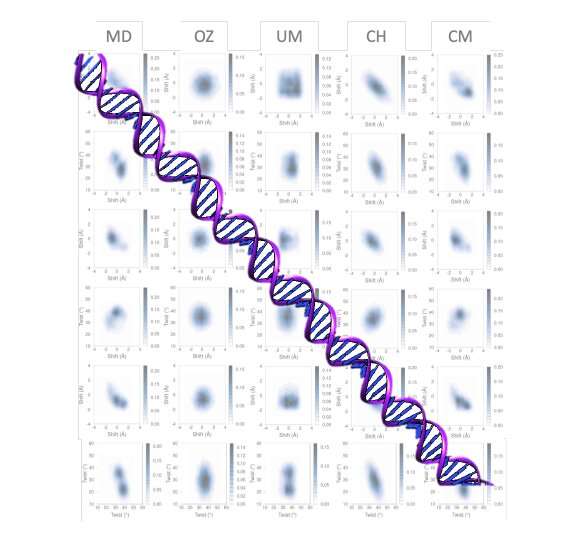
In both physics and chemistry, the mesoscopic scale refers to the length scale on which the properties of a material or phenomenon can be studied, without entering into a discussion about the behavior of individual atoms. In a mesoscopic model, atomic scales are merged with the continuous scale, so they are quite difficult to develop.
A new model of DNA flexibility has been developed by Kim López-Güell, a Maths4Life student, together with Dr. Federica Battistini, under the supervision of Dr. Modesto Orozco in the Molecular Modeling and Bioinformatics laboratory at IRB Barcelona. Using a low-cost computer program, the model developed, which considers the correlation and multimodality at tetramer level, provides results of unprecedented quality.
The model is characterized by its precision and efficiency at the computational level, thereby making it an alternative approach to explore the dynamics of long DNA segments and opening up the possibility of getting closer to the chromatin scale.
“This work is a milestone in the mesoscopic simulation of DNA. It presents a systematic and comprehensive study of DNA movement correlations and a new method to capture them,” says Dr. Battistini, a postdoctoral researcher at IRB Barcelona.
Performed in collaboration with the “BioExcel” Center of Excellence for Computational Biomolecular Research, this work provides a greater understanding of sequencing-dependent DNA at the base pair resolution level. A variety of approaches and simplifications have been used to study this topic over decades but have failed to achieve a multimodal model. The method developed allows a local and global description with high precision for atomic-level molecular simulations and experimental measurements.
DNA movement as an axis
Molecular dynamics is a computational technique that allows simulation of the movement of DNA, its dimeric, trimeric, or tetrameric folding, and even its interaction with proteins and drugs. This technique enables scientists to study processes that occur on time scales ranging from picoseconds to minutes and that apply to molecular systems of various sizes and are therefore pivotal for research into cell functions and disease mechanisms.
This study clarifies how DNA movement works using an approach with a low computational cost that can predict the flexibility and conformation of long DNA strands, which could be extended to RNA duplexes and potentially long polymers. This new model is expected to benefit the scientific community working with nucleic acid simulations.
The findings are published in the journal Nucleic Acids Research.
More information:
Kim López-Güell et al, Correlated motions in DNA: beyond base-pair step models of DNA flexibility, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad136
Provided by
Institute for Research in Biomedicine (IRB Barcelona)
Citation:
A new model predicts the flexibility of DNA movement at the molecular scale (2023, March 31)
retrieved 31 March 2023
from https://phys.org/news/2023-03-flexibility-dna-movement-molecular-scale.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.