The human body contains more than 30 trillion cells. Until recently, the sheer number of cells in the organism meant that approaches to understanding human diseases and developmental processes based on the analysis of single cells were a futuristic vision. The development of new sequencing methods is currently revolutionizing our understanding of cellular heterogeneity. These technologies can detect rare or even new cell types by extracting and sequencing the genetic information from the cells based on ribonucleic acid chains.
In cooperation with Helmholtz Munich, Professor Matthias Meier from the Centre for Biotechnology and Biomedicine at Leipzig University and his research group have developed a new, effective and comparatively inexpensive method to make rare cell types, cell communication types and disease patterns visible in tissue. The researchers have now published their findings in the journal Nature Communications.
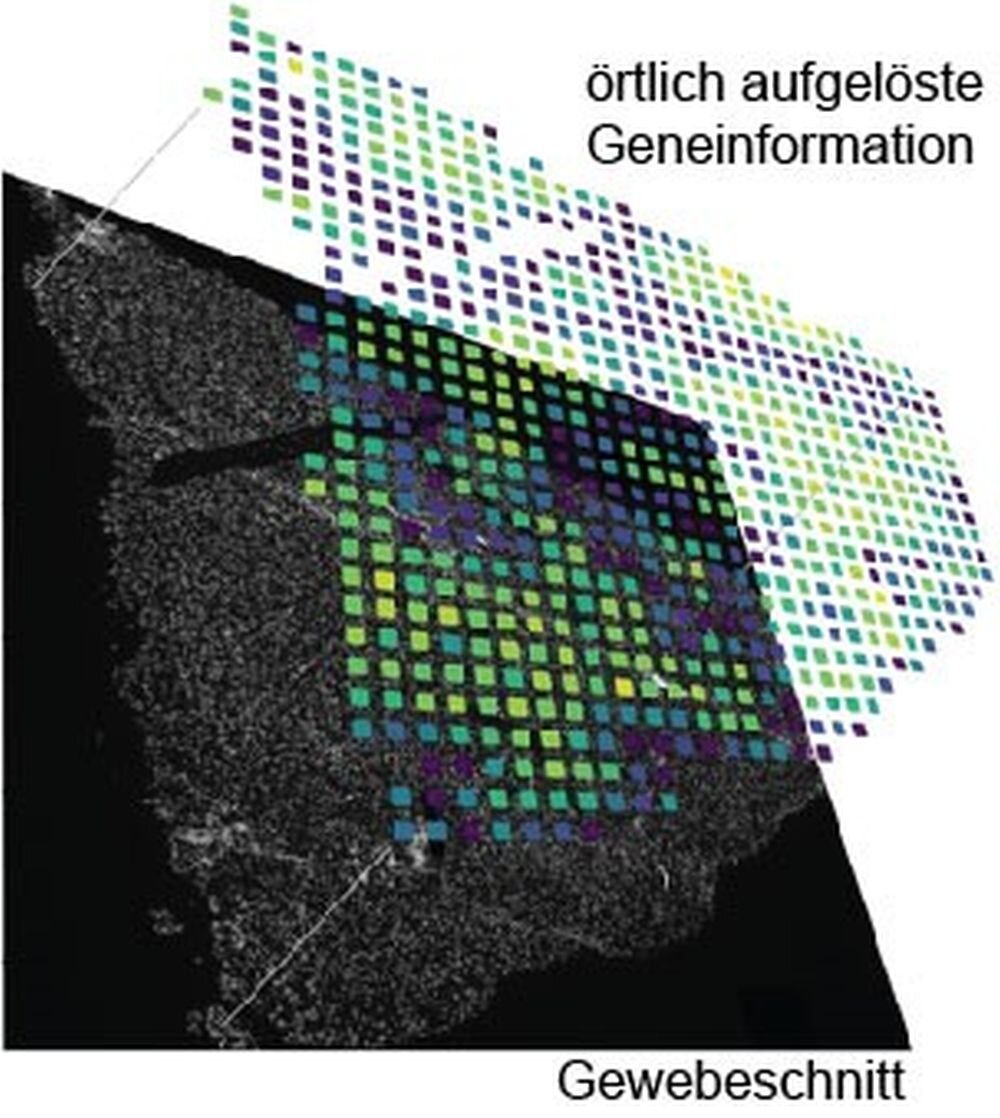
All methods of single-cell analysis require cells to be detached from the tissue composite, losing spatial information about cell types and thus information about the cellular environment, cellular communication pathways or function. To obtain spatially resolved information about individual cells, imaging and sequencing techniques must be used in combination. In recent years, several approaches have been developed to unify the merging of imaging and sequencing data.
Depending on the research question, different parameters such as spatial resolution, detection limit, accessibility of the ribonucleic acids and cost were weighed against each other. An earlier analysis method was based on the idea of attaching local information to the ribonucleic acids using a barcode based on the sequence of DNA bases. After extraction of all the ribonucleic acids and subsequent mass sequencing, the barcodes can be used to create an artificial image.
This is where Johannes Wirth’s work came in. As a doctoral researcher in Matthias Meier’s lab, the researcher at Helmholtz Munich has developed an advanced workflow that makes it possible to acquire locally resolved genomic data paired with high-quality microscopy images. This enables the visualization of rare cell types, cell communication types and disease patterns in tissue. The focus was on the development of a new microfluidic chip that makes it possible to analyze ribonucleic acid chains in large tissue sections at low cost.
“Compared to the original method, the new approach has increased the amount of image information per pixel by a factor of six or twelve. This means that we can resolve about 5000 genes per pixel, which allows us to visualize rare cell types in the kidney or liver,” explains Wirth. By comparison, a standard HD screen can only display the three primary colors with 256 different brightness levels per pixel.
In addition to the technical advances, the team also provided an open source analysis pipeline to make the method easily accessible. As the method is suitable for a wide range of tissues, it will facilitate studies of complex diseases and multi-organ functions and dysfunctions.
“The method we have developed, which combines imaging and sequencing techniques, was a vision until recently. It has revolutionized our understanding of cellular heterogeneity and allowed us to find new cell types in all organisms,” says Professor Meier. With the development of single-cell sequencing methods, it is now possible to better understand cellular developmental pathways and how diseases progress.
More information:
Johannes Wirth et al, Spatial transcriptomics using multiplexed deterministic barcoding in tissue, Nature Communications (2023). DOI: 10.1038/s41467-023-37111-w
Provided by
Leipzig University
Citation:
Making rare cell types visible: Researchers are developing a new method (2023, March 31)
retrieved 31 March 2023
from https://phys.org/news/2023-03-rare-cell-visible-method.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.