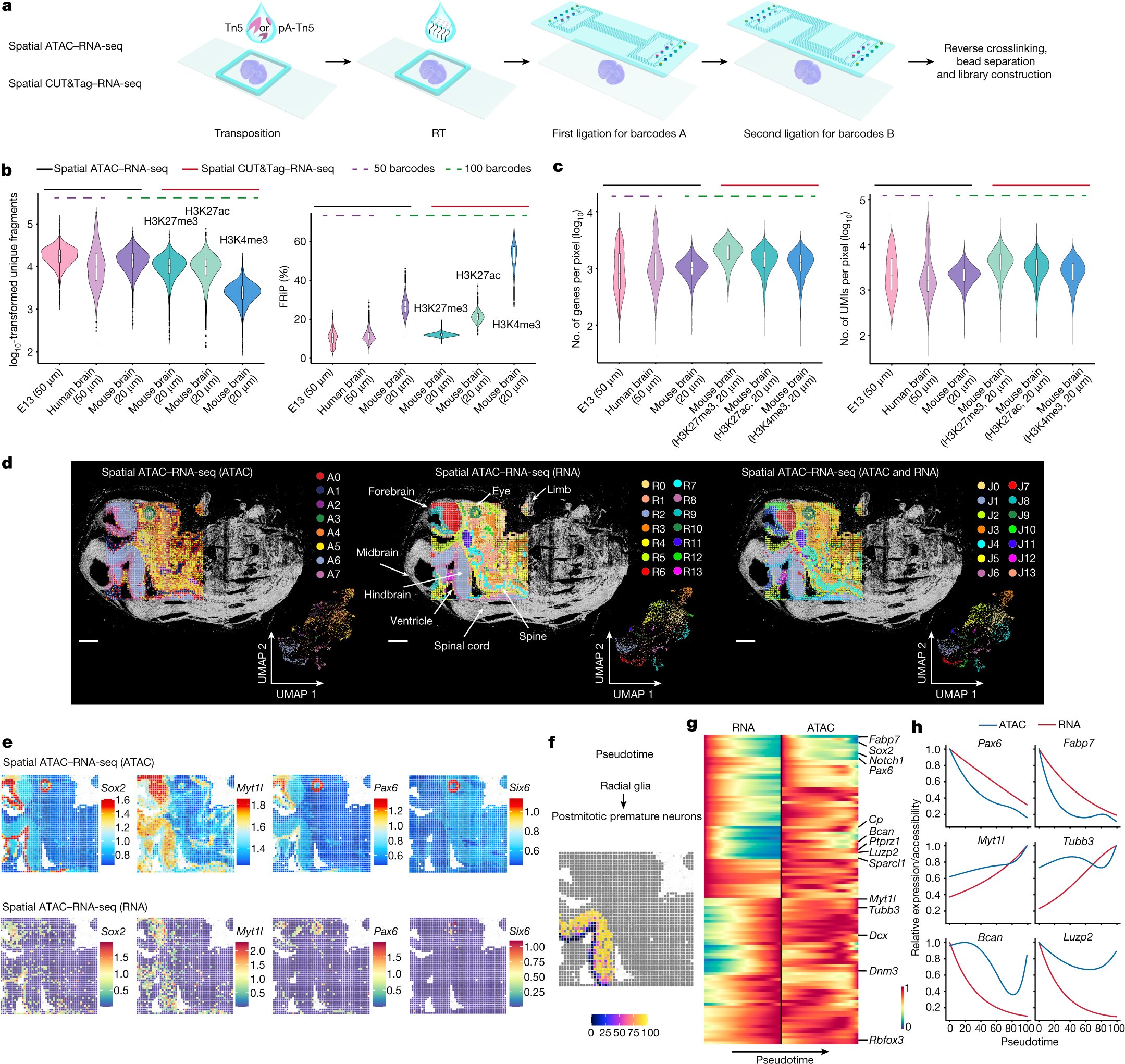
A new study published in Nature reports that a technology known as spatial omics can be used to map simultaneously how genes are switched on and off and how they are expressed in different areas of tissues and organs. This improved technology, developed by researchers at Yale University and Karolinska Institutet, could shed light on the development of tissues, as well as on certain diseases and how to treat them.
Almost all cells in the body have the same set of genes and can in principle become any kind of cell. What distinguishes the cells is how the genes in our DNA are used. In recent years, spatial omics have given us a deeper understanding of how cells read the genome in precise locations in tissues. Now, researchers have further evolved this technology to increase knowledge of how tissues develop and how different diseases arise.
A key part of the study is the researchers’ ability to spatially map simultaneously two crucial components of our genetic makeup, the epigenome and the transcriptome. The epigenome controls the switching mechanisms that turn genes on and off in individual cells, whereas the transcriptome is the result of those gene expressions and what makes each cell unique.
Can detect input and output simultaneously
The epigenome can be thought of as a fuse box. You can flip the switches, but if you can’t see whether the lights are turning on, your information is limited. By spatially mapping both the epigenome and transcriptome, the researchers developed a technology in which both the input (switching on or off a gene) and the output (gene expression) can be detected in the same tissue section. This new technology brings unprecedented insights into gene regulation in precise locations in a tissue.
For this study, the researchers adapted and combined their previously developed techniques to map the epigenome and the transcriptome, and applied these techniques to mouse brains and human brain tissue.
“Now that we can combine the two, we can see both the mechanisms of how the genes are switched on and off, as well as the result,” says Gonçalo Castelo-Branco, professor at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet, and one of the corresponding authors. “This has led us to some unexpected observations, which gives us further insights into how these processes are regulated in different tissue areas and contribute to different cell fates.”
Advancing the field of personalized medicine
The work could bring researchers closer to understanding potential genetic targets for drug therapy, and help advance the field of personalized medicine.
“In the future with this technology, we will be able to really understand in every single patient how those cancer-promoting genes and tumor suppressor genes are being regulated by the epigenetic mechanisms,” says Rong Fan, professor at Yale University and last author of the paper. “The whole epigenetic therapeutics field is just emerging, but I think our technology can potentially empower epigenetic drug discovery.”
More information:
Di Zhang et al, Spatial epigenome–transcriptome co-profiling of mammalian tissues, Nature (2023). DOI: 10.1038/s41586-023-05795-1
Provided by
Karolinska Institutet
Citation:
New technology maps where and how cells read their genome (2023, March 15)
retrieved 16 March 2023
from https://phys.org/news/2023-03-technology-cells-genome.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.

