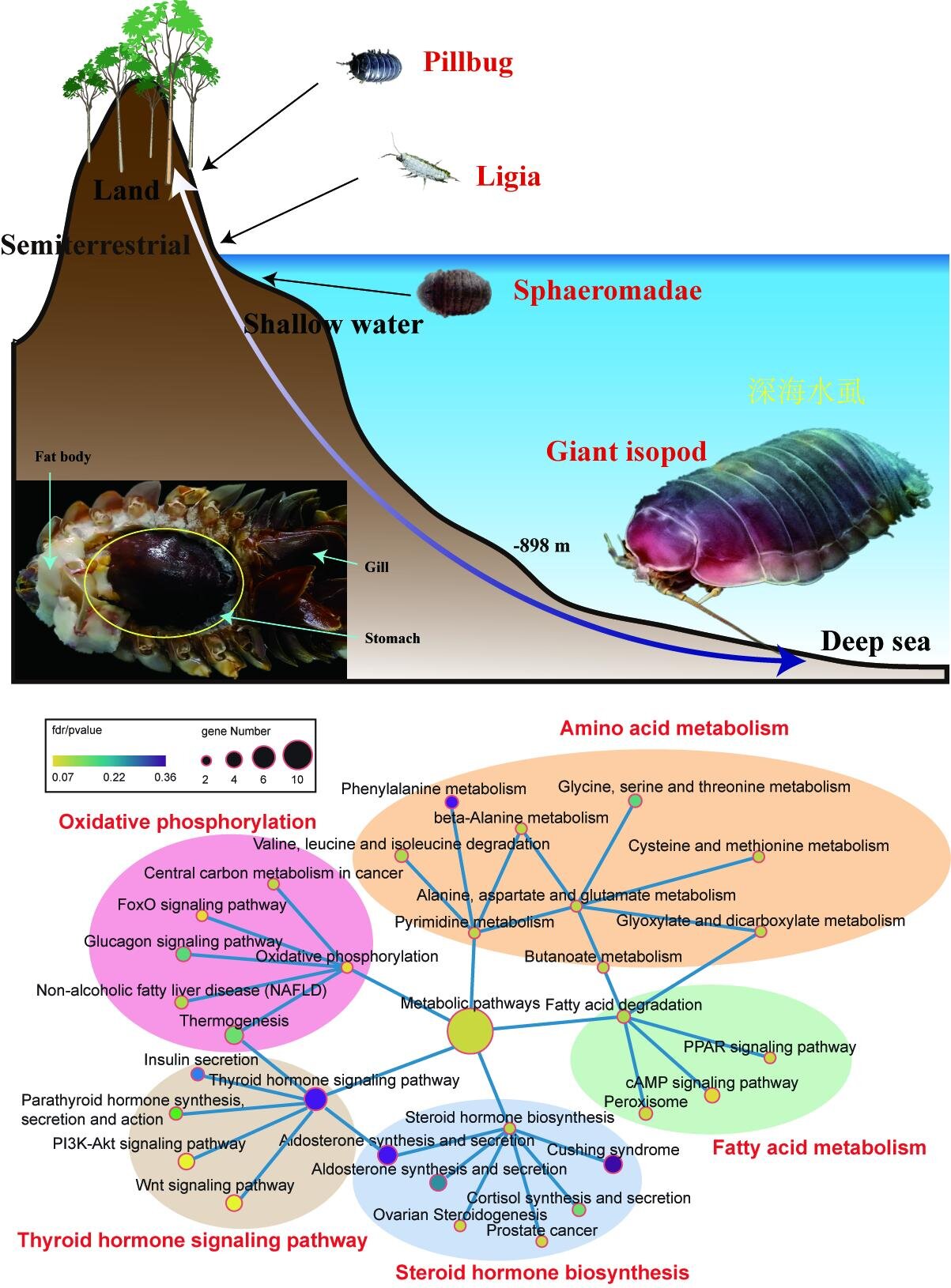
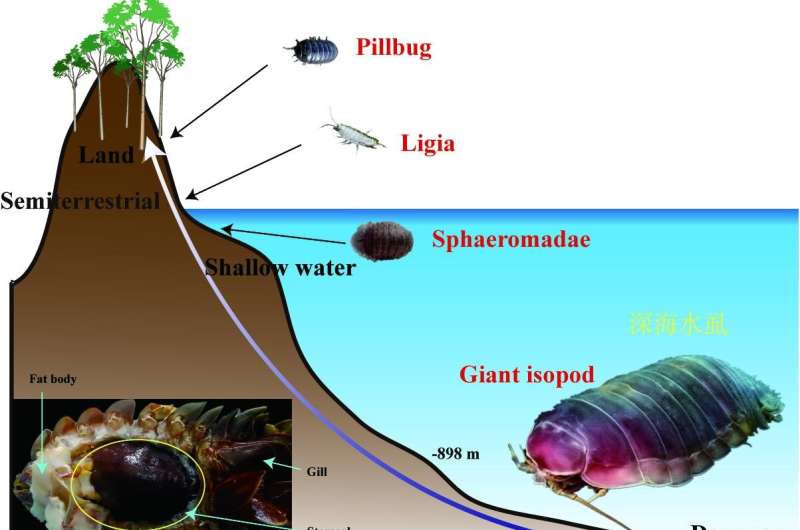
The deep-sea environment is characterized by darkness, low temperature, high hydrostatic pressure and lack of food. Despite the hostile environment, a growing number of deep-dwelling animals have been identified in this ecosystem, including worms, mollusks, fish and crustaceans.
Numerous genomes, including that of tubeworms, mollusks and fishes, have been published in the past few years, whereas no crustacean genome has been reported.
Recently, a research team led by Prof. Li Fuhua and Li Xinzheng from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS) reported the first deep-sea crustacean genome, and illustrated the mechanisms related to body gigantism of deep-sea isopods and specific mechanisms for crustaceans adapting deep-sea environments.
The study was published in BMC Biology on May 13.
The researchers sequenced and assembled a high-quality genome of a giant isopod (Bathynomus jamesi), which has not only large body size, but also large genome size (the largest among sequenced crustaceans), both of which seem to be tightly associated with their adaptation to the deep-sea environment.
Unlike its relatives with small body size, the growth-related pathways of B. jamesi are mostly enriched by expanded gene families, including two hormone signaling pathways (thyroid and insulin). They form a strengthened network of growth-related genes and thus potentially contribute to its body gigantism.
To adapt to the deep-sea oligotrophic environment, B. jamesi has low basal metabolic rate, bulk food storage, and expanded gene families related to nutrient utilization. “The well-developed fat body of B. jamesi is used to store organic reserves, and our results indicate that the lipid accumulation in the fat body should result from low efficiency of lipid degradation rather than high efficiency of lipid synthesis,” said Yuan Jianbo, first author of the study.
The B. jamesi genome can help us understand the evolutionary and migration history (between deep-sea and shallow-water and land) of isopods and even of crustaceans. “This genome also help us understand the genetics related to body gigantism, and uncover the mysterious genetics related to its extraordinary long fasting state, i.e., 5 years’ starvation, and the specific mechanisms for crustaceans adapting [to] deep-sea environments,” said Yuan.
New insights into lipid metabolism associated with pathogenesis of preeclampsia
Jianbo Yuan et al, Genome of a giant isopod, Bathynomus jamesi, provides insights into body size evolution and adaptation to deep-sea environment, BMC Biology (2022). DOI: 10.1186/s12915-022-01302-6
Chinese Academy of Sciences
Citation:
Study reveals the first deep-sea crustacean genome (2022, June 23)
retrieved 23 June 2022
from https://phys.org/news/2022-06-reveals-deep-sea-crustacean-genome.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.

