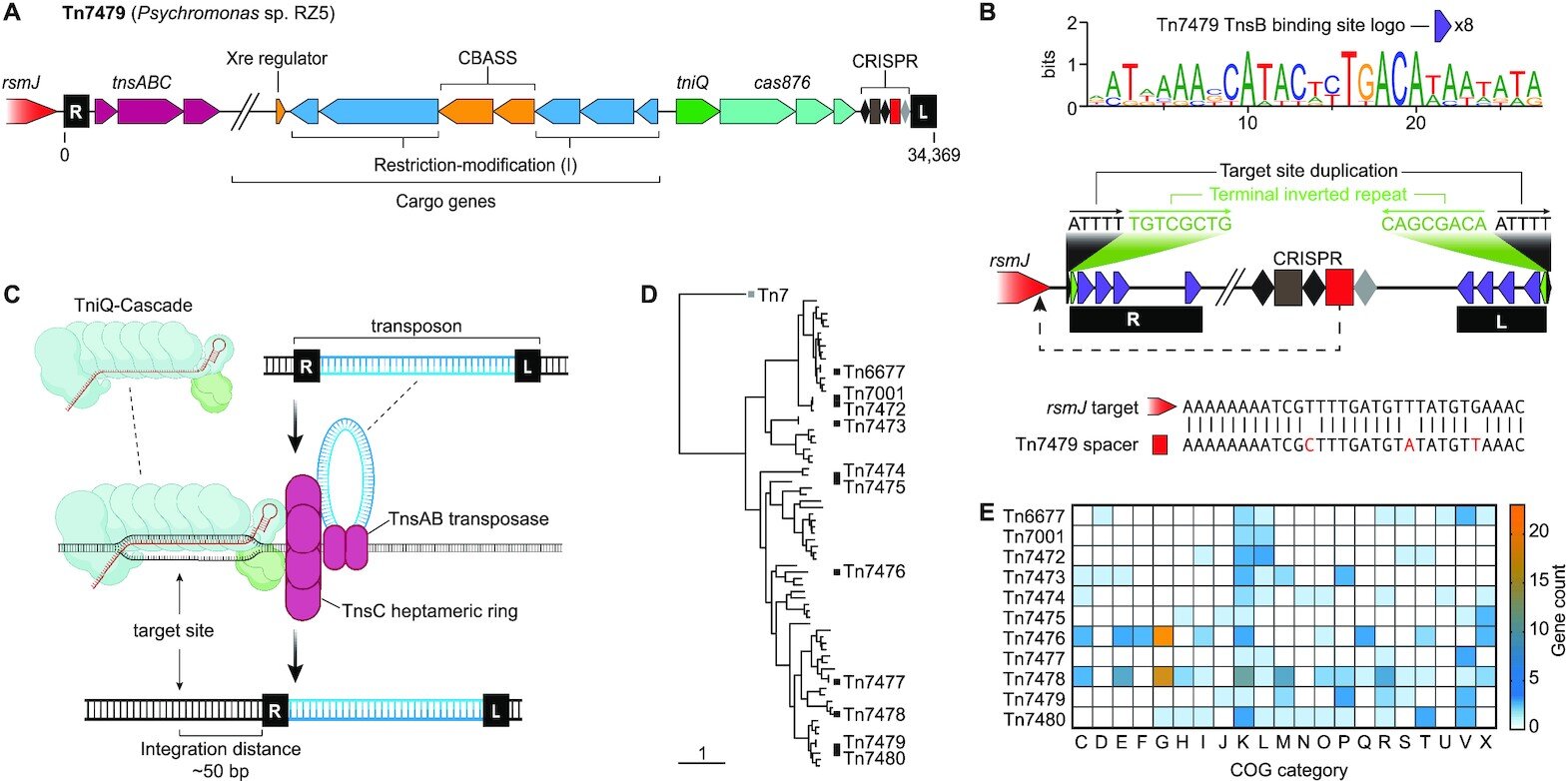
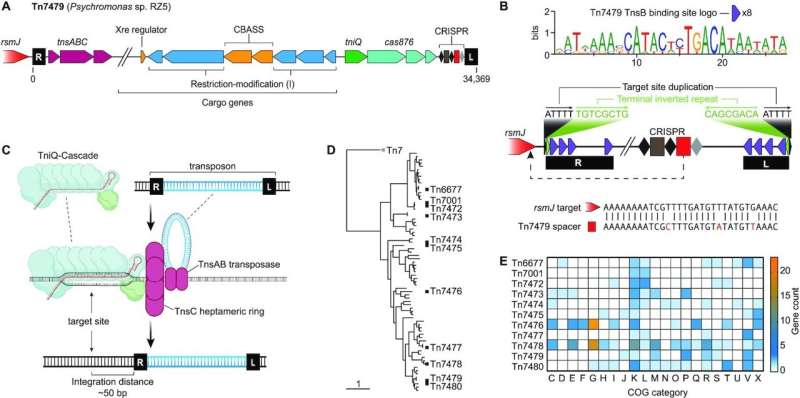
In a new study, North Carolina State University researchers characterize a range of molecular tools to rewrite—not just edit—large chunks of an organism’s DNA, based on CRISPR-Cas systems associated with selfish genetic “hitchhikers” called transposons.
The researchers investigate diverse Type I-F CRISPR-Cas systems and engineer them to add genetic cargo—up to 10,000 additional genetic code letters—to the transposon’s cargo to make desired changes to a bacterium—in this case, E. coli. The paper appears in Nucleic Acids Research.
The findings expand the CRISPR toolbox and could have significant implications in the manipulation of bacteria and other organisms at a time of need for flexible genome editing in therapeutics, biotechnology and more sustainable and efficient agriculture.
Bacteria use CRISPR-Cas as adaptive immune systems to withstand attacks from enemies like viruses. These systems have been adapted by scientists to remove or cut and replace specific genetic code sequences in a variety of organisms. The new finding shows that exponentially larger amounts of genetic code can be moved or added, potentially increasing CRISPR’s functionality.
“In nature, transposons have co-opted CRISPR systems to selfishly move themselves around an organism’s genome to help themselves survive. We’re in turn co-opting what occurs in nature by integrating with transposons a programmable CRISPR-Cas system that can move around genetic cargo that we design to perform some function,” said Rodolphe Barrangou, the Todd R. Klaenhammer Distinguished Professor of Food, Bioprocessing and Nutrition Sciences at NC State and corresponding author of a paper describing the research.
“Using this method, we showed that we can engineer genomes by moving chunks of DNA up to 10,000 letters,” Barrangou said. “Nature already does this—the bioinformatic data shows examples of up to 100,000 genetic letters moved around by transposon-based CRISPR systems—but now we can control and engineer it by using this system.
“To complete the hitchhiking analogy, we’re engineering the hitchhiker to bring certain luggage or cargo into the car to deliver some type of payload when the car arrives at its destination.”
The study shows the researchers proving the method’s effectiveness both in vitro on the lab bench and in vivo in E. coli. The researchers selected 10 different CRISPR-associated transposons to test the method’s effectiveness. The approach worked with all 10 transposons, although they varied in effectiveness based on factors like temperature and the size of the transposon’s cargo load.
“It was exciting to find that all of the systems we tested were functional after reconstructing them into genome editing tools from their native biological forms,” said Avery Roberts, an NC State graduate student and first author of the study. “We uncovered new features of these systems, but there’s likely many more relevant findings and applications to come as the field moves at a rapid pace.”
The research also showed that the method could be used with different transposons at the same time.
“Instead of just one gene—as is the case with other CRISPR systems like the more familiar Type II Cas-9 system—we can bring in a whole metabolic pathway to incorporate a whole new set of functions to an organism,” Barrangou said. “In the future, that could mean providing more flexible disease resistance or drought resistance to plants, for example.”
“We are excited about these findings and see the potential for applying these newly discovered systems in crop plants to accelerate the development of more resilient, higher-yielding varieties,” said Gusui Wu, global head of seeds research for Syngenta Seeds.
Barrangou and Wu add that the work in this study provides a great example of public-private partnerships that drive scientific discovery and train the workforce of tomorrow.
Co-authors of the paper include NC State graduate student Avery Roberts and former NC State Ph.D. student Matthew Nethery.
More information:
Avery Roberts et al, Functional characterization of diverse type I-F CRISPR-associated transposons, Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac985
Provided by
North Carolina State University
Citation:
Genetic ‘hitchhikers’ can be directed using CRISPR (2022, November 21)
retrieved 22 November 2022
from https://phys.org/news/2022-11-genetic-hitchhikers-crispr.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.