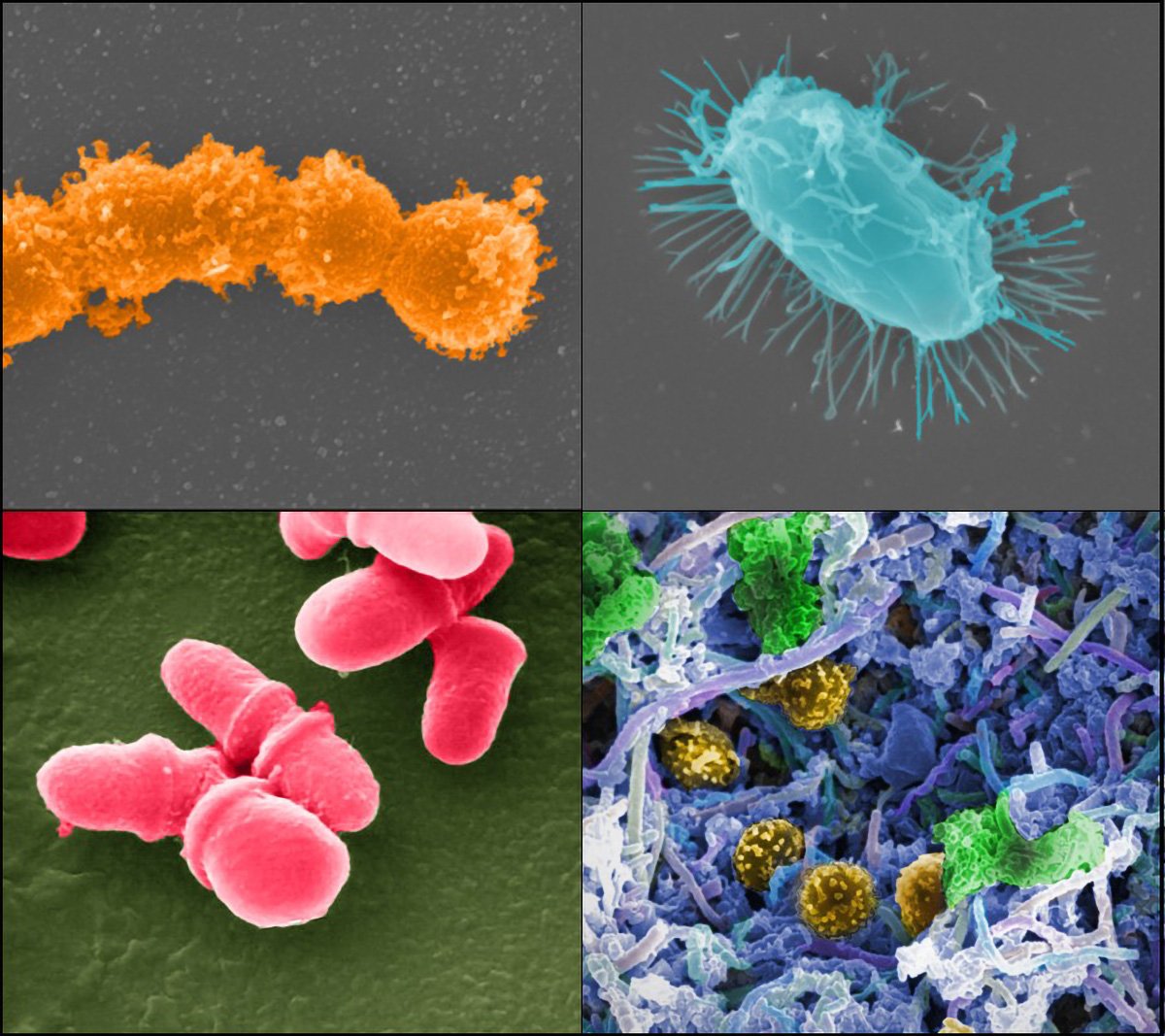
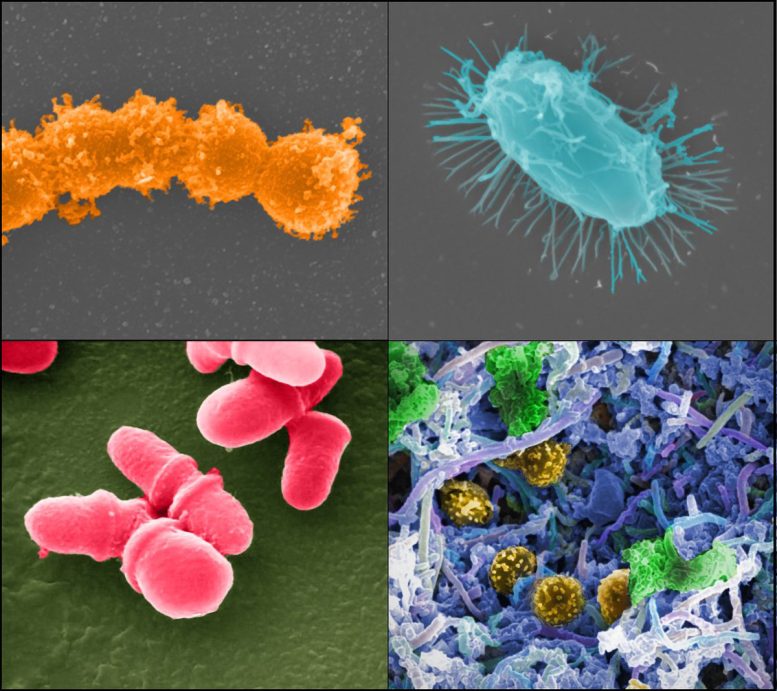
New research reveals that microbes in urban environments are evolving to withstand the very cleaning agents designed to eliminate them. The study also uncovers new strains in Hong Kong, previously only found in the Antarctic desert soil.
Since the recent pandemic, our use of disinfectants has surged, but could our push for sterile urban environments be backfiring?
A new study published in the journal Microbiome has identified novel strains of microbes that have adapted to use the limited resources available in cities and shown that our everyday behavior is changing the makeup of microorganisms in indoor environments.
“Built environments offer distinct conditions that set them apart from natural and engineered habitats,” says Dr. Xinzhao Tong, an assistant professor at Xi’an Jiaotong-Liverpool University (XJTLU), China, and lead author of the study.
“Areas with many buildings are low in the traditional nutrients and essential resources microbes need for survival, so these built environments have a unique microbiome.
“Our use of cleaning and other manufactured products creates a unique setting that puts selective pressures on microbes, which they must adapt to or be eliminated, but the mechanisms by which microbes adapt and survive in built environments are poorly understood,” Dr. Tong explains.
City dwellers
The researchers collected 738 samples from a variety of built environments, including subways, residences, public facilities, piers, and human skin in Hong Kong. They then used shotgun metagenomic sequencing to analyze the microbes’ genomic content and understand how they have adapted to the challenging urban conditions.
The team identified 363 microbial strains that have not been previously identified that live on our skin and the environment around us. Some of these strains’ genomes contained genes for metabolizing manufactured products found in cities and using them as carbon and energy sources. This includes the discovery of a strain of Candidatus phylum Eremiobacterota, previously only reported in Antarctic desert soil.
Specialized metabolic capacities of microbes in oligotrophic built environments. Tong et al. “Diverse and specialized metabolic capabilities of microbes in oligotrophic built environments.” Microbiome (2024). Credit: AJE Video Bytes, in partnership with Springer Nature Group
Dr Tong says: “The genome of this novel strain of Eremiobacterota enables it to metabolize ammonium ions found in cleaning products. The strain also has genes for alcohol and aldehyde dehydrogenases to break down residual alcohol found in common disinfectants.
“Microbes possessing enhanced capabilities to utilize limited resources and tolerate manufactured products, such as disinfectants and metals, out-compete non-resistant strains, enhancing their survival and even evolution within built environments. They could, therefore, pose health risks if they are pathogenic.”
The team identified 11 unique, previously uncharacterized strains of Micrococcus luteus, typically non-pathogenic but capable of causing opportunistic infections in immunocompromised individuals.
“The issue of their adaptation to our behavior becomes particularly critical in clinical settings where hospitals serve as hotspots for diverse pathogens that cause hospital-acquired infections (HAIs). HAIs pose a significant threat, particularly in intensive care units where mortality rates can reach up to 30%,” says Dr Tong.
A balancing act
The researchers also characterized two novel strains of Patescibacteria, known as “nanobacteria”, as they have tiny genomes that do not contain many genes for producing their own resources.
Dr Tong says: “Some strains of Patescibacteria are considered parasitic as they rely on bacterial hosts to supply their nutrients. However, in this study, the researchers found that one of the nanobacteria strains, recovered from human skin, contains genes for the biosynthesis of carotenoids and ubiquinone. These antioxidant compounds are vital to humans, and we typically acquire them, especially carotenoids, through our diets, suggesting a possible mutualistic relationship between bacteria and us as their hosts.”
This enhanced understanding of microbial metabolic functions within built environments helps develop strategies to create a healthy indoor ecosystem of microbes for us to live alongside.
The team is now investigating the transmission and evolution of resistance in pathogenic microbes in intensive care units that are exposed to stringent and extensive disinfectant practices. They hope to improve infection control practices and increase the safety of clinical environments for healthcare workers and patients.
Reference: “Diverse and specialized metabolic capabilities of microbes in oligotrophic built environments” by Xinzhao Tong, Danli Luo, Marcus H. Y. Leung, Justin Y. Y. Lee, Zhiyong Shen, Wengyao Jiang, Christopher E. Mason and Patrick K. H. Lee, 17 October 2024, Microbiome.
DOI: 10.1186/s40168-024-01926-6
Funding: Hong Kong Research Grants Council Research Impact 642 Fund, Natural Science Foundation of Jiangsu Province, Hong Kong Research Grants Council General Research Fund