

Discoveries about the end-replication problem indicate both telomerase and the CST–Polα-primase complex are essential for chromosome protection, suggesting a revision in the science of telomeres and potential impacts on genetic disorders. Credit: SciTechDaily.com
Recent research challenges the long-standing understanding of the end-replication problem in DNA, revealing two distinct issues rather than one.
Half a century ago, scientists Jim Watson and Alexey Olovnikov independently realized that there was a problem with how our DNA gets copied. A quirk of linear DNA replication dictated that telomeres that protect the ends of chromosomes should have been growing shorter with each round of replication, a phenomenon known as the end-replication problem.
Telomerase: A Solution Emerges
But a solution was forthcoming: Liz Blackburn and Carol Greider discovered telomerase, an enzyme that adds the telomeric repeats to the ends of chromosomes. “Case closed, everybody thought,” says Rockefeller’s Titia de Lange.
Now, new research published in Nature suggests that there are two end-replication problems, not one. Further, telomerase is only part of the solution—cells also use the CST–Polα-primase complex, which has been extensively studied in de Lange’s laboratory. “For many decades we thought we knew what the end-replication problem was and how it was solved by telomerase,” says de Lange. “It turns out we had missed half the problem.”
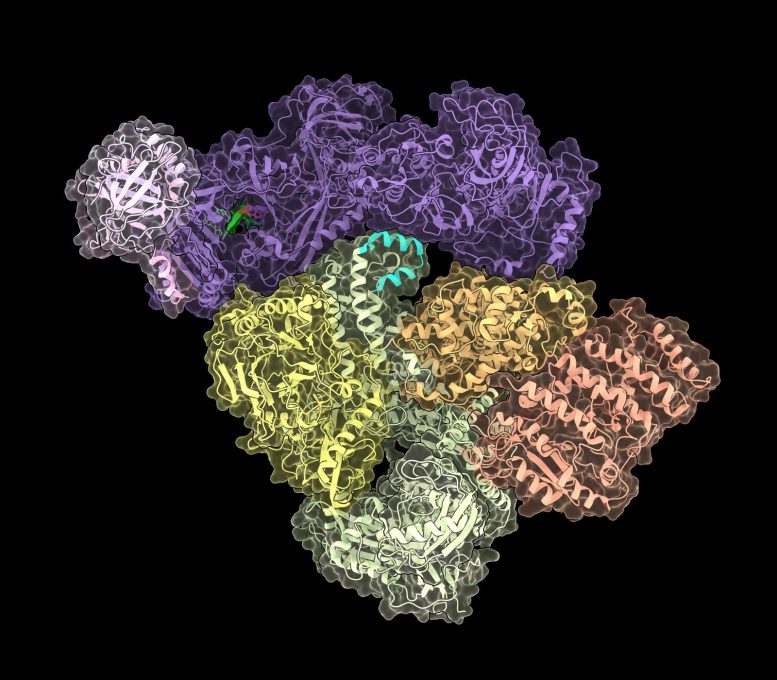
CST–Polα/primase, the enzyme that solves the newly discovered end-replication problem. Credit: Sarah Cai
The Leading-Strand Problem
Since the description of the DNA double helix, it is known that DNA has two complementary strands running in opposite directions—one from 5′ to 3′; the other from 3′ to 5′. When DNA is replicated, the two strands are separated by the replication machinery, also called the replisome. The replisome copies the 3′ to 5′ strand without interruption, a process referred to as leading-strand synthesis. But the other strand is synthesized in short backward steps from many fragments (Okazaki fragments) that are later stitched together.
The process is fairly direct until the ends of the chromosomes. When copying the telomere, leading-strand DNA replication should copy the CCCTAA repeats to generate the TTAGGG repeat strand, while lagging-strand synthesis should do the opposite, making new CCCTAA repeats. The end-replication problem arises because leading strand synthesis fails to reproduce the last part of the telomere, leaving a blunt leading-end telomere without it characteristic and crucial 3’ overhang. Telomerase solves this problem by adding single-stranded TTAGGG repeats to the telomere end. As for the lagging-strand, DNA synthesis should not have a problem. It could start the last Okazaki fragment somewhere along the 3’ overhang.
“The DNA replication machinery cannot fully duplicate the end of a linear DNA, much the same way that you can’t paint the floor under your feet,” says Hiro Takai, senior staff scientist in the de Lange lab and lead author on the paper.
CST–Polα/primase, the enzyme that solves the newly discovered end-replication problem. Credit: Sarah Cai
The Lagging-Strand Problem
As descriptions of biological processes go, this model looked watertight. Until Takai made a surprising discovery while working on cells that lacked molecular machinery called the CST–Polα-primase complex. He and others had previously shown that CST–Polα-primase can replenish CCCTAA repeats at telomeres that had been attacked by DNA-degrading enzymes known as nucleases. This new data revealed something unexpected: not only was the leading strand in need of help—he found evidence that the end of the lagging strand could also not be synthesized by the replisome.
Takai’s work suggested that the end-replication problem was twice as serious as previously thought, impacting both strands of DNA. “The results just didn’t fit with the model for telomere replication,” de Lange says. “At that point, Hiro and I realized that either his results were not right or the model was wrong. As his results seemed very solid to me, we needed to revisit the model.”
De Lange contacted Joseph T. P. Yeeles, a biochemist who studies DNA replication at the Laboratory of Molecular Biology in Cambridge (the same lab where Watson and Crick worked on the structure of the DNA double helix). Yeeles agreed that it would be good to take a close look at how the replisome behaves at the end of a linear DNA template. Could the replisome use a 3’ overhang to make the last Okazaki fragment, as was proposed?
The results of Yeeles’ in vitro replication experiments were very clear. The replisome does not generate Okazaki fragments on the 3’ overhang; it actually stops lagging-strand synthesis long before the leading strand reaches the 5’ end. This second end-replication problem means that both strands of DNA will shorten with each division. Telomerase was only preventing this from happening at the leading strand and Hiro’s data suggested that CST–Polα-primase fixed the second end-replication problem, that of the lagging strand.
Takai spent the next four years designing new assays to confirm Yeeles’ findings in vivo. He was able to measure how much DNA is lost due to the lagging-strand end-replication problem, revealing how many CCCAAT repeats need to be added by CST–Polα-primase to keep telomeres intact.
Implications and Future Directions
The results change our understanding of telomere biology—requiring revision of the textbooks. But the findings may also have clinical implications. Individuals who inherit mutations in CST–Polα-primase suffer from telomere disorders, such as Coats plus syndrome, which is characterized by an eye disorder and abnormalities in the brain, bones, and GI tract. Through a better understanding of how we maintain our telomeres, strides could one day be made in addressing these devastating disorders.
Reference: “Cryo-EM structure of the human CST–Polα/primase complex in a recruitment state” by Sarah W. Cai, John C. Zinder, Vladimir Svetlov, Martin W. Bush, Evgeny Nudler, Thomas Walz and Titia de Lange, 16 May 2022, Nature Structural & Molecular Biology.
DOI: 10.1038/s41594-022-00766-y

