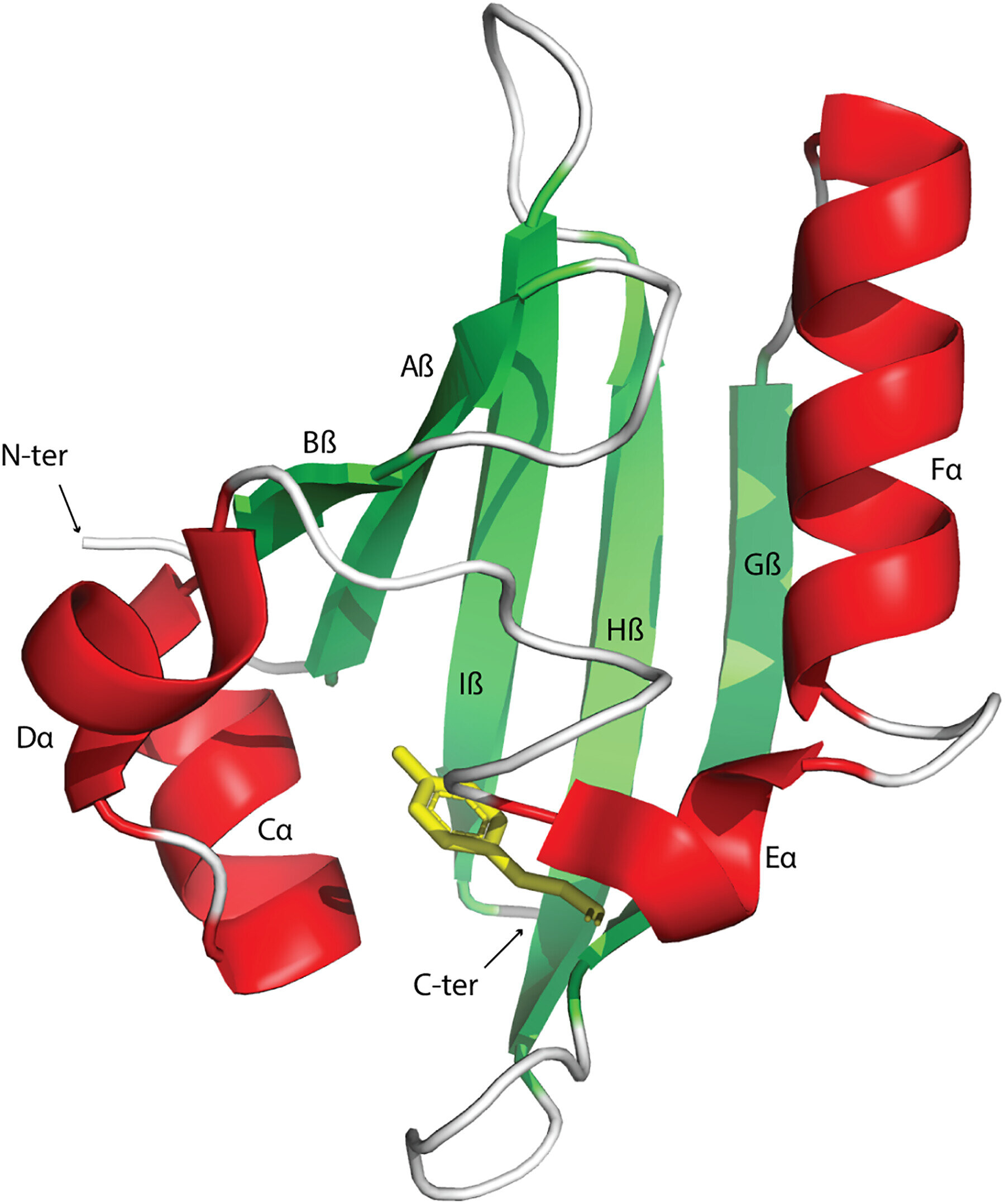
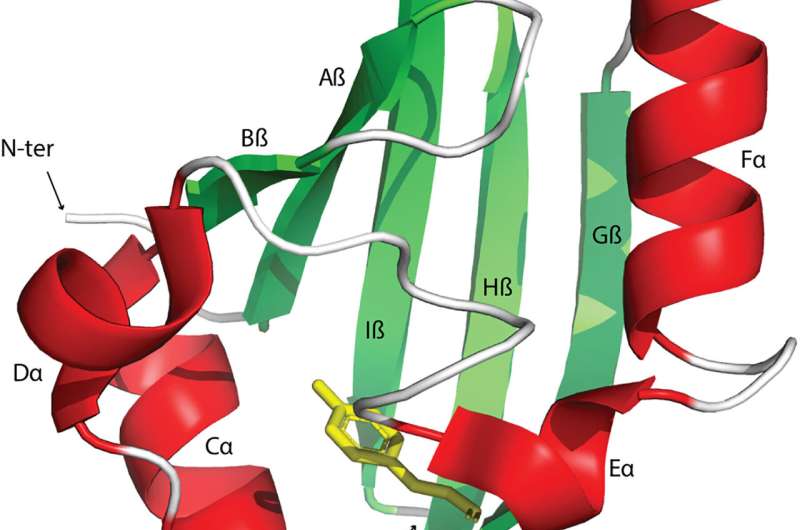
Signal transduction and perception regulates biological activities to adapt to changing environments. The Pert-Arnt-Sim domains are commonly available sensors found across diverse receptors in bacteria, eukaryotes, and archaea. However, the extent of their functional diversity and their distribution across the tree of life remains to be characterized.
In a new report in Science Advances, Jiawei Xing and a team of scientists in biology and translational data at the Ohio State University, U.S., have used sequence conservation and structural information to propose specific and potential functions for nearly three million Pert-Arnt-Sim domains (abbreviated as PAS domains). The work suggested the origin of these domains in bacteria and their horizontal transfer to archaea and eukaryotes. The close relationship of the domains between humans and bacteria provides a unique opportunity to drive drug design.
The influence of sensor domains on biological pathways
Signal transduction pathways are central avenues in all living cells that detect nutrients, hormones, oxygen, and redox potential to regulate a variety of cellular functions. These signals can be recognized by receptor proteins by using dedicated sensor domains. The PAS domains are found in transcription factors, protein kinases, and enzymes regulating messenger turnover and chemoreceptors. Although sensor domains are typically extracytoplasmic, these domains are mainly cytoplasmic. Moreover, the PAS domains present in human transcription factors are key drug targets for cancer therapy.
In this work, Xing et al. provided an extensive comparative genomic analysis of PAS domains across bacteria, archaea, and eukaryotes. The team showed the origins of PAS domains in bacteria and how their sensory functions evolved through different paths. The eukaryotes attained PAS domains from bacteria through a variety of independent gene transfer events.
The similarity of human PAS domains to those in bacteria suggest the functionality of these proteins as useful models to determine signal specificity relative to human counterparts. These outcomes pave the way to explore functional studies and drug development in PAS domains, while serving as a framework to study protein families with diverse and versatile functions.
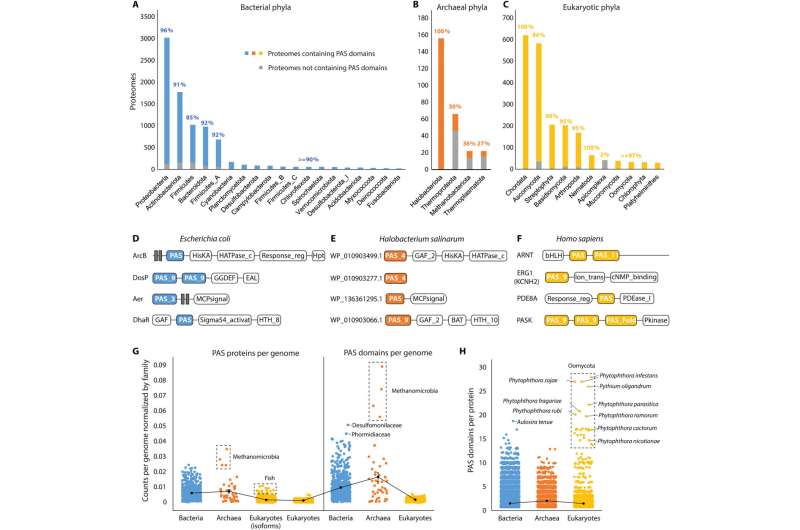
PAS domain distribution across the tree of life
Since originally discovering the PAS superfamily, the distributions of the domains remain to be systematically investigated across the tree of life. Xing and colleagues searched for these domains in the entire set of reference proteomes to establish the presence of the domain in 66% of archaeal, 93% of bacterial and 93% of eukaryotic proteomes. The domains were widely present in bacterial phyla, while unevenly distributed in archaea and widely distributed in eukaryotes.
The scientists collected all PAS-containing proteins from the InterPro database and identified their domain composition. While histidine kinases are the most common PAS proteins in bacteria and archaea, transcription factors are more available in eukaryotes. The archaeal and bacterial genomes on average encoded more of the proteins and domains than eukaryotic genomes.
The existing classification of PAS domains do not reflect their biological functions
The present pfam database of protein families classifies the PAS domain into 17 families. To explore if the present classification of PAS domains reflect their biological function, Xing et al. conducted a comprehensive literature search. The work showed that the existing classification of PAS domains did not reflect sensory functions.
The team also conducted a BLAST search using sequences from families. They followed up these experiments by identifying PAS families using the sequence and structural information from the NCBI reference sequence database, as well as the entire pfam database.
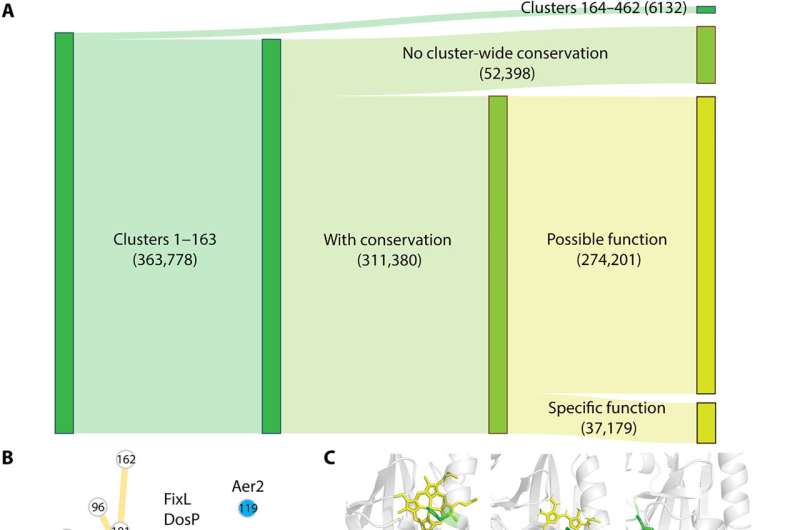
Key findings
Xing and colleagues showed that these domains have short and diverse sequences that complicated their phylogenetic analysis. However, the domains shared the same structural fold. The team sampled 1% of sequences from each cluster and constructed phylogenetic trees via a structure-guided approach for short and diverse sequences.
They noted the sequence, structural and phylogenetic information to suggest that PAS domains in such clusters independently evolved the capabilities for heme-binding. The study also revealed the eukaryotic PAS domains to have bacterial origins, alongside a capacity to offer promising drug targets.
Outlook
In this way, the research team conducted a genome-wide survey of the PAS (Pert-Arnt-Sim) domains 24 years after the most recent analysis. In this work, the team analyzed nearly 3 million PAS domains from large-scale genomes of bacteria, archaea, and eukaryotes. The researchers presented the findings about evolution, the genomic landscape, and functional diversification of biological sensors. The outcomes showed several PAS domains to be present in a variety of life forms, while also identifying them in major types of signal transduction proteins.
Since the existing classification systems are outdated to an extent, Xing and team used several methods to produce a better classification system in this work. The study revealed the PAS domains as universal molecular sensors to design targeted experiments for validation and further study.
More information:
Jiawei Xing et al, Origin and functional diversification of PAS domain, a ubiquitous intracellular sensor, Science Advances (2023). DOI: 10.1126/sciadv.adi4517
Gregg L. Semenza, Hypoxia-Inducible Factors in Physiology and Medicine, Cell (2012). DOI: 10.1016/j.cell.2012.01.021
© 2023 Science X Network
Citation:
The origin and functional diversification of the Pert-Arnt-Sim (PAS) domain—an intracellular sensor (2023, September 6)
retrieved 6 September 2023
from https://phys.org/news/2023-09-functional-diversification-pert-arnt-sim-pas-domainan.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.