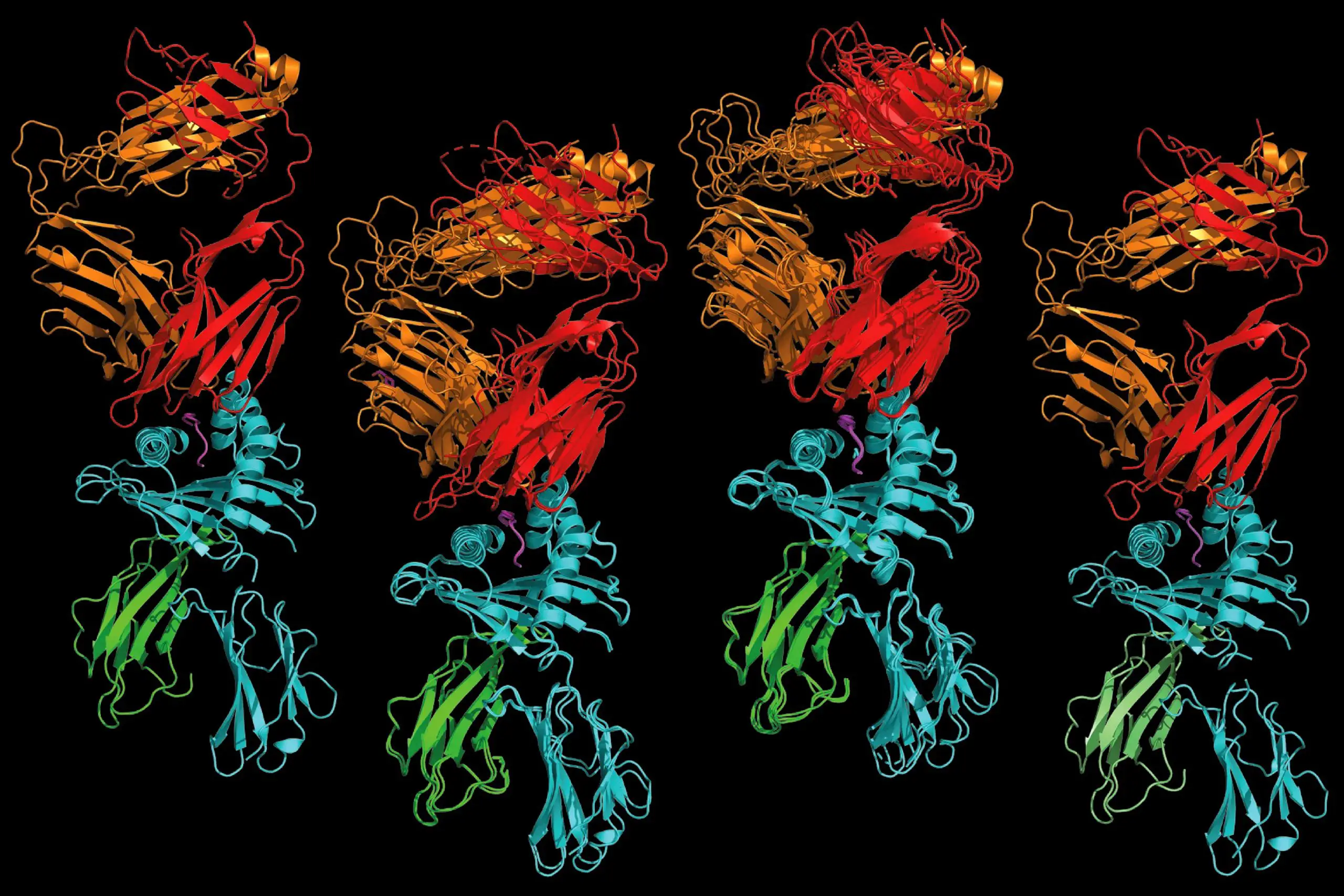

A T cell receptor that recognizes a human protein fragment (left) is remarkably similar to one that recognizes a bacterial protein fragment (right), and to two receptors capable of recognizing both human and bacterial protein fragments (middle). A study by researchers at Washington University School of Medicine in St. Louis and colleagues at Stanford University and Oxford University supports the idea that some T cells that react to microbes also may react to normal human proteins, causing autoimmune disease. The findings promise to accelerate efforts to improve diagnostic tools and treatments for autoimmune diseases. Credit: Xinbo Yang and K. Chris Garcia
Study opens pathway to better diagnosis and treatment of autoimmune diseases.
Researchers from Washington University, Stanford University, and Oxford University have developed a method to identify protein fragments and immune cells that cause autoimmune reactions. The study focuses on the HLA-B*27 variant linked to ankylosing spondylitis and acute anterior uveitis. The breakthrough could lead to improved diagnosis and treatment of autoimmune diseases.
Autoimmune diseases are thought to be the result of mistaken identity. Immune cells on patrol, armed and ready to defend the body against invading pathogens, mistake normal human cells for infected cells and turn their weapons on their own healthy tissues. In most cases, though, finding the source of the confusion — the tiny fragment of normal human protein that looks dangerously similar to a protein from a pathogen — has been challenging for scientists. That missing piece of the puzzle has hampered efforts to develop effective diagnostics and specific therapies for many autoimmune conditions.
That finally may be changing. A team involving researchers from Washington University School of Medicine in St. Louis, Stanford University School of Medicine, and Oxford University has developed a way to find crucial protein fragments that drive autoimmunity, as well as the immune cells that respond to them. The findings, published in the journal Nature, open a promising pathway to diagnose and treat autoimmune diseases.
“Of all genes, the HLA genes have the greatest amount of variation across the human population. There are many, many autoimmune diseases that are associated with specific variants of the HLA genes, and in most cases, we don’t know why,” said co-senior author Wayne M. Yokoyama, MD, the Sam J. Levin and Audrey Loew Levin Professor of Arthritis Research at Washington University. “This paper outlines a strategy for figuring out why certain HLA variants are linked to certain diseases. It also provides strong evidence that cross-reactivity between human and microbial proteins drives autoimmunity in at least two diseases and probably many others. Now that we understand the underlying drivers, we can start focusing on the approaches that are most likely to yield benefits for patients.”
The autoimmune diseases ankylosing spondylitis, which involves arthritis in the spine and pelvis, and acute anterior uveitis, which is characterized by inflammation in the eye, are both strongly associated with an HLA variant called HLA-B*27. The link between ankylosing spondylitis and HLA-B*27 was discovered 50 years ago — making it one of the first such associations identified between disease and HLA variants — and it remains one of the strongest known associations between any disease and an HLA variant.
The HLA family of proteins is involved in helping immune cells detect invading pathogens and distinguishing between microbial and human proteins, and is highly variable across individuals. HLA proteins function like hands that pick up fragments of whichever proteins are lying about — microbial or human — and show them to immune cells called T cells to figure out if they’re a sign of danger (microbial) or not (human).
T cells don’t recognize protein fragments by themselves; they recognize the fragment plus the hand that holds it. Scientists have long assumed that the combination of this particular hand — HLA-B*27 — plus a bit of an unknown human protein was being misidentified as dangerous in people with either of the two diseases, triggering autoimmune attacks in the eye or the spine. But for decades, they couldn’t find the fragment. Some scientists began to speculate that the misidentification hypothesis was wrong and some other reason accounted for the association between HLA-B*27 and the two diseases.
Co-corresponding author K. Christopher Garcia, PhD, and co-first author Xinbo Yang, PhD, of Stanford Medicine, along with co-corresponding authors Geraldine M. Gillespie, PhD, and Andrew J. McMichael, PhD, and co-first author Lee Garner, PhD, of Oxford University, collaborated with Yokoyama and co-first author Michael Paley, MD, PhD, of Washington University on a novel way to find the elusive fragment. The research team identified certain T cells that were abundant in the blood and joints of people with ankylosing spondylitis, and in the eyes of people with uveitis.
Garcia and Yang then devised a way to identify protein fragments that drive a T cell response when combined with HLA-B*27, and mapped the fragments against the human genome and five bacterial genomes to identify proteins from which the fragments may have originated. Using that approach, they were able to narrow down the millions of possibilities to a very short list of human and microbial proteins. Then, they determined the structures of the detector molecules — known as T cell receptors — on T cells from both groups of patients and compared them. The similarities were striking.
“This study reveals the power of studying T cell specificity and activity from the ground up; that is, identifying the T cells that are most active in a given response, followed by identifying what they respond to,” Garcia said. “Clearly these patient-derived TCRs are seeing a spectrum of common antigens, and that may be driving the autoimmunity. Proving this in humans is very difficult, but that is our future direction and could lead to therapeutics.”
The findings reveal key aspects of the biological mechanisms underlying ankylosing spondylitis, anterior uveitis and potentially many other autoimmune diseases.
“By combining recently developed technologies, we have revisited an old hypothesis that asks if the traditional antigen-presenting function of HLA-B*27 contributes to disease initiation or pathogenesis in the autoimmune conditions ankylosing spondylitis and uveitis,” Gillespie said. “Our findings that T cells at the sites of pathology recognize HLA-B*27 bound to both self and microbial antigens adds a very important layer of understanding to these complex conditions that also feature strong inflammatory signatures. Our hope is that this work will one day pave the way for more targeted therapies, not only for these conditions but ultimately, for other autoimmune diseases.”
By providing strong support for the idea that T cells that react to microbes also may react to normal human proteins, the findings promise to accelerate efforts to improve diagnostic tools and treatments for autoimmune diseases.
“For ankylosing spondylitis, the average time between initial symptoms and actual diagnosis is seven to eight years,” said Paley, an assistant professor of medicine, of ophthalmology, and of pathology & immunology. “Shortening that time with improved diagnostics could make a dramatic impact on patients’ lives, because treatment could be initiated earlier. As for therapeutics, if we could target these disease-causing T cells for elimination, we could potentially cure a patient or maybe even prevent the disease in people with the high-risk genetic variant. There’s a lot of potential for clinical benefit here.”
Reference: “Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides” by Xinbo Yang, Lee I. Garner, Ivan V. Zvyagin, Michael A. Paley, Ekaterina A. Komech, Kevin M. Jude, Xiang Zhao, Ricardo A. Fernandes, Lynn M. Hassman, Grace L. Paley, Christina S. Savvides, Simon Brackenridge, Max N. Quastel, Dmitriy M. Chudakov, Paul Bowness, Wayne M. Yokoyama, Andrew J. McMichael, Geraldine M. Gillespie and K. Christopher Garcia, 7 December 2022, Nature.
DOI: 10.1038/s41586-022-05501-7
Funding: NIH/National Institutes of Health, Howard Hughes Medical Institute, Medical Research Council, Oxford University McMichael Trust Fund, Rosetrees Trust, Oxford University John Fell Fund, Oxford University Medical Sciences Division, NIHR Oxford Biomedical Research Centre, Ministry of Science and Higher Education of the Russian Federation, Rheumatology Research Foundation, Arthritis National Research Foundation, Rheumatic Diseases Research Resource-Based Center, Andrew M. and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs, Foundation for Barnes-Jewish Hospital